Learning Objectives
Following this assignment students should be able to:
- Explain the similarities and differences between a file and a directory.
- Translate an absolute path into a relative path and vice versa.
- Construct absolute and relative paths that identify specific files and directories.
- Identify the actual command, flags, and filenames in a command-line call.
- Demonstrate the use of tab completion, and explain its advantages.
Reading
Lecture Notes
1) Week 2 “Quiz” on Canvas
NOTE: Question 13 will have you upload your code for the below activity!
2) Summary Sheets Due Thursday
This is a sheet of paper where you use a writing instrument to create a mindmap / cheat sheet.
Exercises
-- Navigating the Commandline --
Navigating the computer from the command line
In this exercise we will be following a Software Carpentry Lesson. It is suggested that you have this page open to follow along and a bash shell window.
The Unix Shell
credit:

Copyright (c) Software Carpentry
see also
http://swcarpentry.github.io/shell-novice/
Intro
Learning Objectives
- Explain the similarities and differences between a file and a directory.
- Translate an absolute path into a relative path and vice versa.
- Construct absolute and relative paths that identify specific files and directories.
- Identify the actual command, flags, and filenames in a command-line call.
- Demonstrate the use of tab completion, and explain its advantages.
For this we will be using UW JupyterHub Terminal instance.
The file will be preloaded on your instance but from reference this is the file - data-shell.zip
The part of the operating system responsible for managing files and directories is called the file system.ß It organizes our data into files, which hold information, and directories (also called “folders”), which hold files or other directories.
Several commands are frequently used to create, inspect, rename, and delete files and directories. To start exploring them, let’s open a shell window:
$The dollar sign is a prompt, which shows us that the shell is waiting for input; your shell may show something more elaborate.
Type the command
whoami, then press the Enter key (sometimes marked Return) to send the command to the shell. The command’s output is the ID of the current user, i.e., it shows us who the shell thinks we are:$ whoamijovyanMore specifically, when we type
whoamithe shell:- finds a program called
whoami, - runs that program,
- displays that program’s output, then
- displays a new prompt to tell us that it’s ready for more commands.
Next, let’s find out where we are by running a command called
pwd(which stands for “print working directory”). At any moment, our current working directory is our current default directory, i.e., the directory that the computer assumes we want to run commands in unless we explicitly specify something else. Here, the computer’s response is/home/jovyan$ pwd/home/jovyan
To understand what a “home directory” is, let’s have a look at how the file system as a whole is organized. At the top is the root directory that holds everything else. We refer to it using a slash character
/on its own; this is the leading slash in/home/jovyan.Notice that there are two meanings for the
/character. When it appears at the front of a file or directory name, it refers to the root directory. When it appears inside a name, it’s just a separator.
So that we have the same experience lets navigate to the ‘data-shell’ directory.
ls
Let’s see what’s in this directory by running
ls, which stands for “listing”:$ lscreatures molecules pizza.cfg data north-pacific-gyre solar.pdf Desktop notes.txt writing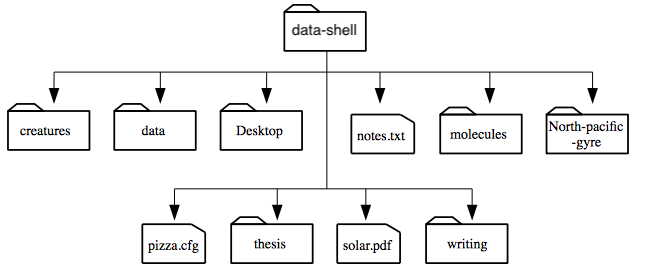
lsprints the names of the files and directories in the current directory in alphabetical order, arranged neatly into columns. We can make its output more comprehensible by using the flag-F, which tellslsto add a trailing/to the names of directories:$ ls -Fcreatures/ molecules/ pizza.cfg data/ north-pacific-gyre/ solar.pdf Desktop/ notes.txt writing/Here, we can see that
data-shellcontains seven sub-directories. The names that don’t have trailing slashes, likenotes.txt,pizza.cfg, andsolar.pdf, are plain old files. And note that there is a space betweenlsand-F: without it, the shell thinks we’re trying to run a command calledls-F, which doesn’t exist.relative path
Now let’s take a look at what’s in
data-shelldirectory by runningls -F data, i.e., the commandlswith the arguments-Fanddata. The second argument — the one without a leading dash — tellslsthat we want a listing of something other than our current working directory:$ ls -F dataamino-acids.txt animal-counts/ animals.txt elements/ morse.txt pdb/ planets.txt salmon.txt sunspot.txtThe output shows us that there are four text files and two sub-sub-directories. Organizing things hierarchically in this way helps us keep track of our work: it’s possible to put hundreds of files in our home directory, just as it’s possible to pile hundreds of printed papers on our desk, but it’s a self-defeating strategy.
Notice, by the way that we spelled the directory name
data. It doesn’t have a trailing slash: that’s added to directory names bylswhen we use the-Fflag to help us tell things apart. And it doesn’t begin with a slash because it’s a relative path, i.e., it tellslshow to find something from where we are, rather than from the root of the file system.absolute path
If we run
ls -F /data(with a leading slash) we get a different answer, because/datais an absolute path:$ ls -F /dataNote you will get an “No file” warning here. This is because we this directory does not exist.
The leading
/tells the computer to follow the path from the root of the filesystem, so it always refers to exactly one directory, no matter where we are when we run the command.If we wanted to use the *absolute path to list out the contents of this directory we could used
$ ls -F /home/jovyan/data-shell/data/Note this would work no matter what our pwd is.
Nelle’s Pipeline: Organizing Files
Knowing just this much about files and directories, Nelle is ready to organize the files that the protein assay machine will create. First, she creates a directory called
north-pacific-gyre(to remind herself where the data came from). Inside that, she creates a directory called2012-07-03, which is the date she started processing the samples. She used to use names likeconference-paperandrevised-results, but she found them hard to understand after a couple of years. (The final straw was when she found herself creating a directory calledrevised-revised-results-3.)Nelle names her directories “year-month-day”, with leading zeroes for months and days, because the shell displays file and directory names in alphabetical order. If she used month names, December would come before July; if she didn’t use leading zeroes, November (‘11’) would come before July (‘7’).
Each of her physical samples is labelled according to her lab’s convention with a unique ten-character ID, such as “NENE01729A”. This is what she used in her collection log to record the location, time, depth, and other characteristics of the sample, so she decides to use it as part of each data file’s name. Since the assay machine’s output is plain text, she will call her files
NENE01729A.txt,NENE01812A.txt, and so on. All 1520 files will go into the same directory.If she is in her home directory, Nelle can see what files she has using the command:
$ ls north-pacific-gyre/2012-07-03/This is a lot to type, but she can let the shell do most of the work. If she types:
$ ls norand then presses tab, the shell automatically completes the directory name for her:
$ ls north-pacific-gyre/If she presses tab again, Bash will add
2012-07-03/to the command, since it’s the only possible completion. Pressing tab again does nothing, since there are 1520 possibilities; pressing tab twice brings up a list of all the files, and so on. This is called tab completion, and we will see it in many other tools as we go on.Key Points
- The file system is responsible for managing information on the disk.
- Information is stored in files, which are stored in directories (folders).
- Directories can also store other directories, which forms a directory tree.
/on its own is the root directory of the whole filesystem.- A relative path specifies a location starting from the current location.
- An absolute path specifies a location from the root of the filesystem.
- Directory names in a path are separated with
/on Unix, but\on Windows. ..means “the directory above the current one”;.on its own means “the current directory”.- Most files’ names are
something.extension. The extension isn’t required, and doesn’t guarantee anything, but is normally used to indicate the type of data in the file. - Most commands take options (flags) which begin with a
-.
Creating Things
Objectives
- Create a directory hierarchy that matches a given diagram.
- Create files in that hierarchy using an editor or by copying and renaming existing files.
- Display the contents of a directory using the command line.
- Delete specified files and/or directories.
We now know how to explore files and directories, but how do we create them in the first place? Let’s go back to
data-shell, and usels -Fto see what it contains:$ ls -F creatures/ molecules/ pizza.cfg data/ north-pacific-gyre/ solar.pdf Desktop/ notes.txt writing/Let’s create a new directory called
thesisusing the commandmkdir thesis(which has no output):$ mkdir thesisAs you might (or might not) guess from its name,
mkdirmeans “make directory”. Sincethesisis a relative path (i.e., doesn’t have a leading slash), the new directory is made below the current working directory:$ ls -F creatures/ north-pacific-gyre/ thesis/ data/ notes.txt writing/ Desktop/ pizza.cfg molecules/ solar.pdfHowever, there’s nothing in it yet:
$ ls -F thesisLet’s change our working directory to
thesisusingcd, then run a text editor called Nano to create a file calleddraft.txt:$ cd thesis $ nano draft.txtWhich Editor?
When we say, “
nanois a text editor,” we really do mean “text”: it can only work with plain character data, not tables, images, or any other human-friendly media. We use it in examples because almost anyone can drive it anywhere without training, but please use something more powerful for real work. On Unix systems (such as Linux and Mac OS X), many programmers use Emacs or Vim (both of which are completely unintuitive, even by Unix standards), or a graphical editor such as Gedit. On Windows, you may wish to use Notepad++. On a Mac, TextWrangler is an option.No matter what editor you use, you will need to know where it searches for and saves files. If you start it from the shell, it will (probably) use your current working directory as its default location. If you use your computer’s start menu, it may want to save files in your desktop or documents directory instead. You can change this by navigating to another directory the first time you “Save As…”
Let’s type in a few lines of text, then use Control-O to write our data to disk:
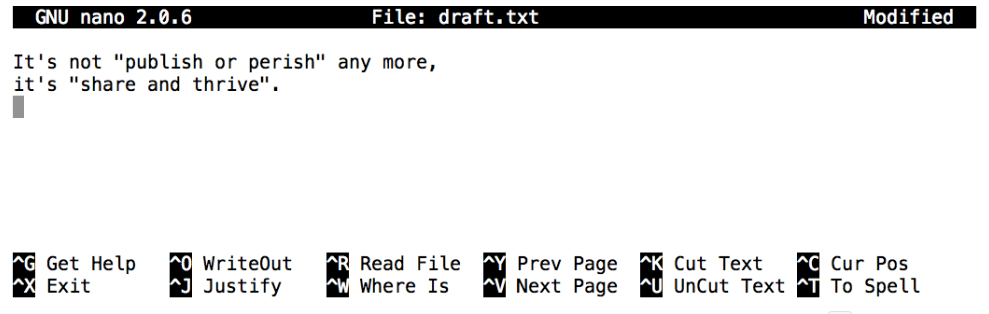
Once our file is saved, we can use Control-X to quit the editor and return to the shell. (Unix documentation often uses the shorthand
^Ato mean “control-A”.)nanodoesn’t leave any output on the screen after it exits, butlsnow shows that we have created a file calleddraft.txt:$ ls draft.txtLet’s tidy up by running
rm draft.txt:$ rm draft.txtThis command removes files (“rm” is short for “remove”). If we run
lsagain, its output is empty once more, which tells us that our file is gone:$ lsDeleting Is Forever
Unix doesn’t have a trash bin: when we delete files, they are unhooked from the file system so that their storage space on disk can be recycled. Tools for finding and recovering deleted files do exist, but there’s no guarantee they’ll work in any particular situation, since the computer may recycle the file’s disk space right away.
Pipes and Filters
Objectives
- Redirect a command’s output to a file.
- Process a file instead of keyboard input using redirection.
- Construct command pipelines with two or more stages.
- Explain what usually happens if a program or pipeline isn’t given any input to process.
- Explain Unix’s “small pieces, loosely joined” philosophy.
Now that we know a few basic commands, we can finally look at the shell’s most powerful feature: the ease with which it lets us combine existing programs in new ways. We’ll start with a directory called
moleculesthat contains six files describing some simple organic molecules. The.pdbextension indicates that these files are in Protein Data Bank format, a simple text format that specifies the type and position of each atom in the molecule.$ ls molecules cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdbword count
Let’s go into that directory with
cdand run the commandwc *.pdb.wcis the “word count” command: it counts the number of lines, words, and characters in files. The*in*.pdbmatches zero or more characters, so the shell turns*.pdbinto a complete list of.pdbfiles:$ cd molecules $ wc *.pdb 20 156 1158 cubane.pdb 12 84 622 ethane.pdb 9 57 422 methane.pdb 30 246 1828 octane.pdb 21 165 1226 pentane.pdb 15 111 825 propane.pdb 107 819 6081 totalWildcards
*is a wildcard. It matches zero or more characters, so*.pdbmatchesethane.pdb,propane.pdb, and so on. On the other hand,p*.pdbonly matchespentane.pdbandpropane.pdb, because the ‘p’ at the front only matches itself.?is also a wildcard, but it only matches a single character. This means thatp?.pdbmatchespi.pdborp5.pdb, but notpropane.pdb. We can use any number of wildcards at a time: for example,p*.p?*matches anything that starts with a ‘p’ and ends with ‘.’, ‘p’, and at least one more character (since the ‘?’ has to match one character, and the final*can match any number of characters). Thus,p*.p?*would matchpreferred.practice, and evenp.pi(since the first*can match no characters at all), but notquality.practice(doesn’t start with ‘p’) orpreferred.p(there isn’t at least one character after the ‘.p’).When the shell sees a wildcard, it expands the wildcard to create a list of matching filenames before running the command that was asked for. This means that commands like
wcandlsnever see the wildcard characters, just what those wildcards matched. This is another example of orthogonal design.If we run
wc -linstead of justwc, the output shows only the number of lines per file:$ wc -l *.pdb 20 cubane.pdb 12 ethane.pdb 9 methane.pdb 30 octane.pdb 21 pentane.pdb 15 propane.pdb 107 totalWe can also use
-wto get only the number of words, or-cto get only the number of characters.redirect
Which of these files is shortest? It’s an easy question to answer when there are only six files, but what if there were 6000? Our first step toward a solution is to run the command:
$ wc -l *.pdb > lengthsThe
>tells the shell to redirect the command’s output to a file instead of printing it to the screen. The shell will create the file if it doesn’t exist, or overwrite the contents of that file if it does. (This is why there is no screen output: everything thatwcwould have printed has gone into the filelengthsinstead.)ls lengthsconfirms that the file exists:$ ls lengths lengthscat
We can now send the content of
lengthsto the screen usingcat lengths.catstands for “concatenate”: it prints the contents of files one after another. There’s only one file in this case, socatjust shows us what it contains:$ cat lengths 20 cubane.pdb 12 ethane.pdb 9 methane.pdb 30 octane.pdb 21 pentane.pdb 15 propane.pdb 107 totalsort
Now let’s use the
sortcommand to sort its contents. We will also use the -n flag to specify that the sort is numerical instead of alphabetical. This does not change the file; instead, it sends the sorted result to the screen:$ sort -n lengths 9 methane.pdb 12 ethane.pdb 15 propane.pdb 20 cubane.pdb 21 pentane.pdb 30 octane.pdb 107 totalhead
We can put the sorted list of lines in another temporary file called
sorted-lengthsby putting> sorted-lengthsafter the command, just as we used> lengthsto put the output ofwcintolengths. Once we’ve done that, we can run another command calledheadto get the first few lines insorted-lengths:$ sort -n lengths > sorted-lengths $ head -1 sorted-lengths 9 methane.pdbUsing the parameter
-1withheadtells it that we only want the first line of the file;-20would get the first 20, and so on. Sincesorted-lengthscontains the lengths of our files ordered from least to greatest, the output ofheadmust be the file with the fewest lines.pipe
If you think this is confusing, you’re in good company: even once you understand what
wc,sort, andheaddo, all those intermediate files make it hard to follow what’s going on. We can make it easier to understand by runningsortandheadtogether:$ sort -n lengths | head -1 9 methane.pdbThe vertical bar between the two commands is called a pipe. It tells the shell that we want to use the output of the command on the left as the input to the command on the right. The computer might create a temporary file if it needs to, or copy data from one program to the other in memory, or something else entirely; we don’t have to know or care.
We can use another pipe to send the output of
wcdirectly tosort, which then sends its output tohead:$ wc -l *.pdb | sort -n | head -1 9 methane.pdbHere’s what actually happens behind the scenes when we create a pipe. When a computer runs a program—any program—it creates a process in memory to hold the program’s software and its current state. Every process has an input channel called standard input. (By this point, you may be surprised that the name is so memorable, but don’t worry: most Unix programmers call it “stdin”. Every process also has a default output channel called standard output (or “stdout”).
The shell is actually just another program. Under normal circumstances, whatever we type on the keyboard is sent to the shell on its standard input, and whatever it produces on standard output is displayed on our screen. When we tell the shell to run a program, it creates a new process and temporarily sends whatever we type on our keyboard to that process’s standard input, and whatever the process sends to standard output to the screen.
Here’s what happens when we run
wc -l *.pdb > lengths. The shell starts by telling the computer to create a new process to run thewcprogram. Since we’ve provided some filenames as parameters,wcreads from them instead of from standard input. And since we’ve used>to redirect output to a file, the shell connects the process’s standard output to that file.If we run
wc -l *.pdb | sort -ninstead, the shell creates two processes (one for each process in the pipe) so thatwcandsortrun simultaneously. The standard output ofwcis fed directly to the standard input ofsort; since there’s no redirection with>,sort’s output goes to the screen. And if we runwc -l *.pdb | sort -n | head -1, we get three processes with data flowing from the files, throughwctosort, and fromsortthroughheadto the screen.This simple idea is why Unix has been so successful. Instead of creating enormous programs that try to do many different things, Unix programmers focus on creating lots of simple tools that each do one job well, and that work well with each other. This programming model is called pipes and filters. We’ve already seen pipes; a filter is a program like
wcorsortthat transforms a stream of input into a stream of output. Almost all of the standard Unix tools can work this way: unless told to do otherwise, they read from standard input, do something with what they’ve read, and write to standard output.The key is that any program that reads lines of text from standard input and writes lines of text to standard output can be combined with every other program that behaves this way as well. You can and should write your programs this way so that you and other people can put those programs into pipes to multiply their power.
Redirecting Input
As well as using
>to redirect a program’s output, we can use<to redirect its input, i.e., to read from a file instead of from standard input. For example, instead of writingwc ammonia.pdb, we could writewc < ammonia.pdb. In the first case,wcgets a command line parameter telling it what file to open. In the second,wcdoesn’t have any command line parameters, so it reads from standard input, but we have told the shell to send the contents ofammonia.pdbtowc’s standard input.Nelle’s Pipeline: Checking Files
Nelle has run her samples through the assay machines and created 1520 files in the
north-pacific-gyre/2012-07-03directory described earlier. As a quick sanity check, she types:$ cd north-pacific-gyre/2012-07-03 $ wc -l *.txtThe output is 1520 lines that look like this:
300 NENE01729A.txt 300 NENE01729B.txt 300 NENE01736A.txt 300 NENE01751A.txt 300 NENE01751B.txt 300 NENE01812A.txt ... ...Now she types this:
$ wc -l *.txt | sort -n | head -5 240 NENE02018B.txt 300 NENE01729A.txt 300 NENE01729B.txt 300 NENE01736A.txt 300 NENE01751A.txtWhoops: one of the files is 60 lines shorter than the others. When she goes back and checks it, she sees that she did that assay at 8:00 on a Monday morning—someone was probably in using the machine on the weekend, and she forgot to reset it. Before re-running that sample, she checks to see if any files have too much data:
$ wc -l *.txt | sort -n | tail -5 300 NENE02040A.txt 300 NENE02040B.txt 300 NENE02040Z.txt 300 NENE02043A.txt 300 NENE02043B.txtThose numbers look good—but what’s that ‘Z’ doing there in the third-to-last line? All of her samples should be marked ‘A’ or ‘B’; by convention, her lab uses ‘Z’ to indicate samples with missing information. To find others like it, she does this:
$ ls *Z.txt NENE01971Z.txt NENE02040Z.txtSure enough, when she checks the log on her laptop, there’s no depth recorded for either of those samples. Since it’s too late to get the information any other way, she must exclude those two files from her analysis. She could just delete them using
rm, but there are actually some analyses she might do later where depth doesn’t matter, so instead, she’ll just be careful later on to select files using the wildcard expression*[AB].txt. As always, the*matches any number of characters; the expression[AB]matches either an ‘A’ or a ‘B’, so this matches all the valid data files she has.
Key Points
command > fileredirects a command’s output to a file.first | secondis a pipeline: the output of the first command is used as the input to the second.- The best way to use the shell is to use pipes to combine simple single-purpose programs (filters).
