Taking Megan for a spin for Renee
Read files
ls /home/shared/16TB_HDD_01/fish546/renee/build
F1B-KM40_merged.fastq.gz
F1B-KM40_trimmed_R1_paired.fastq.gz
F1B-KM40_trimmed_R1_unpaired.fastq.gz
F1B-KM40_trimmed_R2_paired.fastq.gz
F1B-KM40_trimmed_R2_unpaired.fastq.gz
F2R-KM41_merged.fastq.gz
F2R-KM41_trimmed_R1_paired.fastq.gz
F2R-KM41_trimmed_R1_unpaired.fastq.gz
F2R-KM41_trimmed_R2_paired.fastq.gz
F2R-KM41_trimmed_R2_unpaired.fastq.gz
fastp.html
fastp.json
md5checksums_metagenomes_2025.txt
megahit
raw-data
Trimmomatic-0.39Diamond blast
/home/shared/diamond-2.1.8 blastx \
--db /home/shared/16TB_HDD_01/sam/databases/blastdbs/ncbi-nr-20250429.dmnd \
--query /home/shared/16TB_HDD_01/fish546/renee/F2R-KM41_merged.fastq.gz \
--out ../output/sr-blastx.daa \
--outfmt 100 \
--top 5 \
--block-size 15.0 \
--index-chunks 4 \
--threads 32Meganizer
/home/shared/megan-6.24.20/tools/daa-meganizer \
--in ../output/sr-blastx.daa \
--threads 32 \
--mapDB ../data/megan-map-Feb2022.dbdaa2rma
/home/shared/megan-6.24.20/tools/daa2rma \
--in ../output/sr-blastx.daa \
--mapDB ../data/megan-map-Feb2022.db \
--out ../output/sr-blastx-meganized.rma6 \
--threads 40 2>&1 | tee --append ../output/daa2rma_log.txt
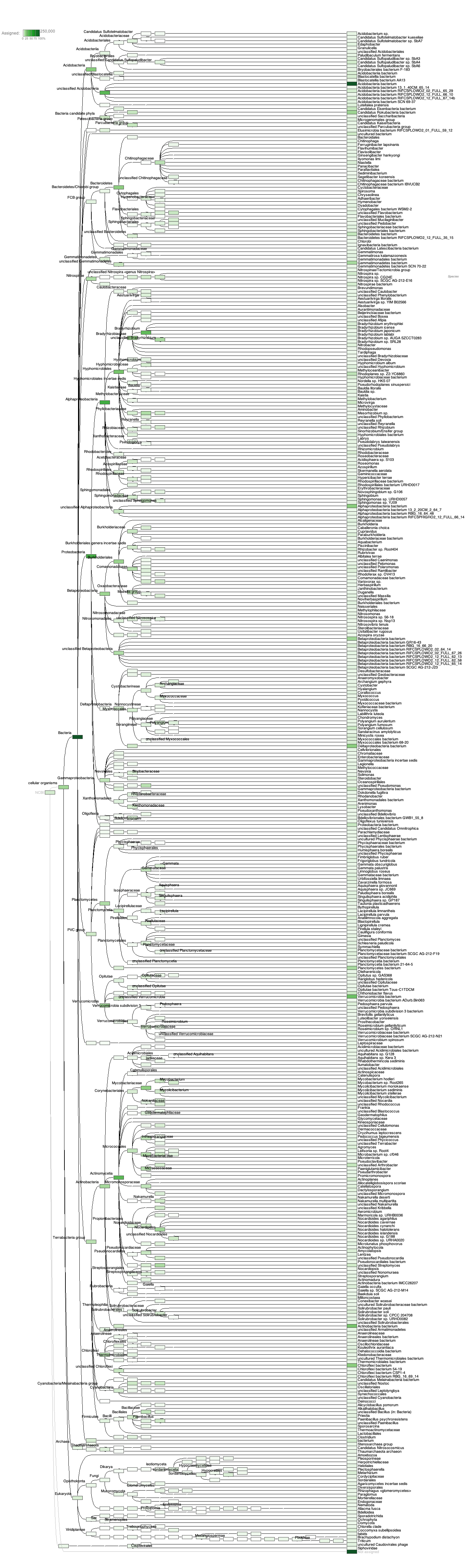
Open MEGAN on locally and looked at rda file
pdf: http://gannet.fish.washington.edu/seashell/snaps/sr-blastx-meganized.pdf