The directory /volume1/web/nightingales/G_macrocephalus contains seven sequencing projects, each representing a distinct Azenta sequencing batch or data delivery. The projects vary in size and file-naming conventions but consistently use paired-end FASTQ/FQ files with clear sample identifiers. Two projects (30-1067895835, 30-1149633765) are earlier batches, while others correspond to newer Azenta runs (AN00025267, AN00025268, H202SC23041287).
- 30-1067895835 — Single-sample pilot sequencing set (
1D11), paired-end reads, includes Azenta report.
- 30-1149633765 — Large-scale sequencing project (
EXP###,MHW###prefixes), paired-end reads with checksums and HTML report.
- 30-1149634506 — Moderate batch (
ET##prefix), paired-end reads with standard naming and report.
- 30-943133806 — Mixed sample identifiers (e.g.,
19-G,RESUB-###), includes supplemental folder and Azenta report.
- AN00025267 / AN00025268 — Consistent naming (
##B_1.fastq.gz/_2.fastq.gz), likely replicate or continuation projects.
- H202SC23041287 — Deeply nested structure (
GM##samples, multiple lanes per run), each sample has multiple files and MD5 checksums.
30-1067895835
30-1067895835 is test aging sample, noting there are also some comparable shark sample(s). refers to WGBS from a single cod fin clip (experimental cod with “genetic sampling count” = 84 from this table - that cod was held at 9C).
| Project | Sample ID | Barcode Sequence | # Reads | Yield (Mbases) | Mean Quality Score | % Bases >= 30 |
|---|---|---|---|---|---|---|
| 30-1067895835 | 1D11 | CAACACAG+TGGCTATC | 47,537,334 | 14,261 | 37.27 | 87.02 |
Direct links - https://owl.fish.washington.edu/nightingales/G_macrocephalus/30-1067895835/1D11_R1_001.fastq.gz - https://owl.fish.washington.edu/nightingales/G_macrocephalus/30-1067895835/1D11_R2_001.fastq.gz
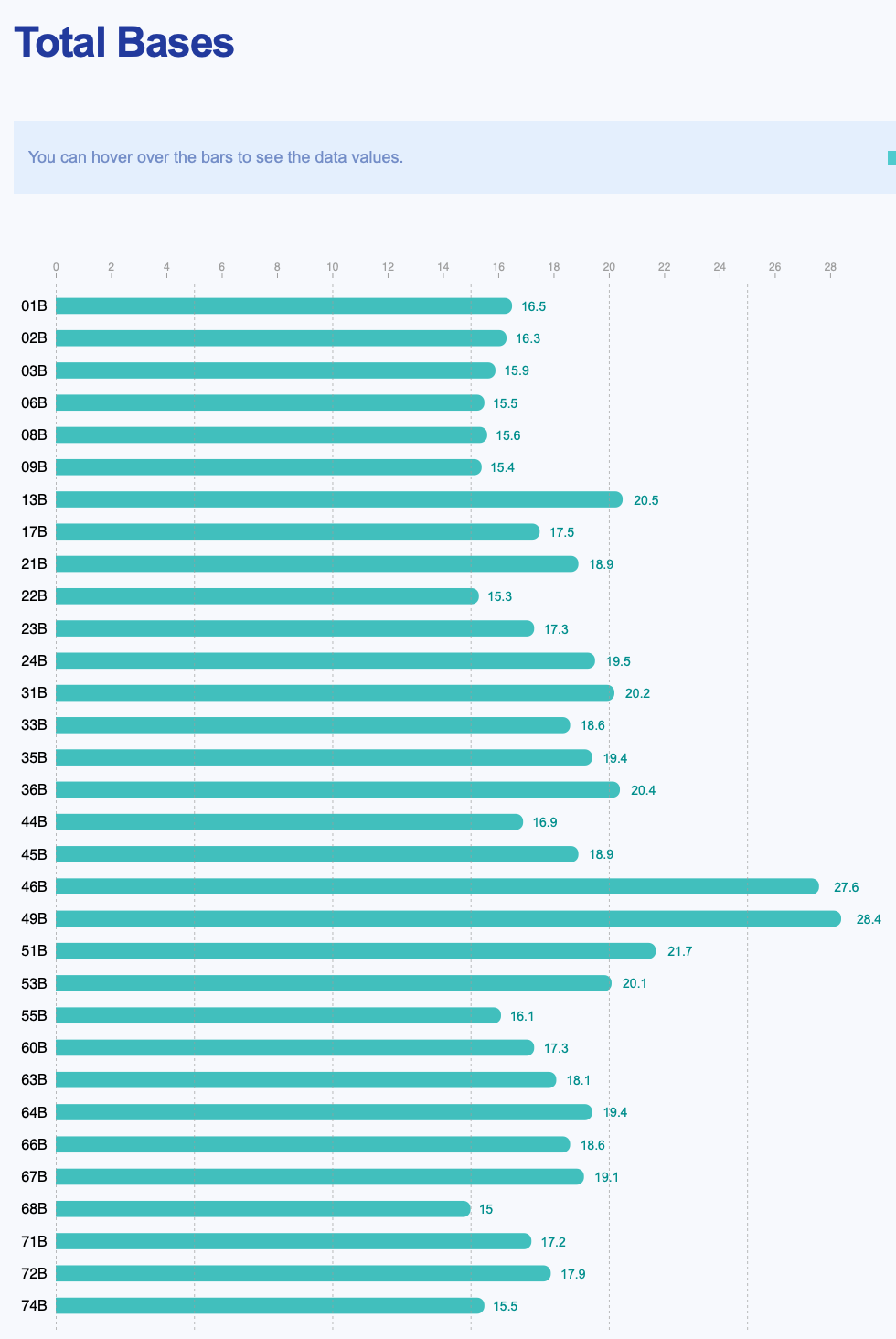
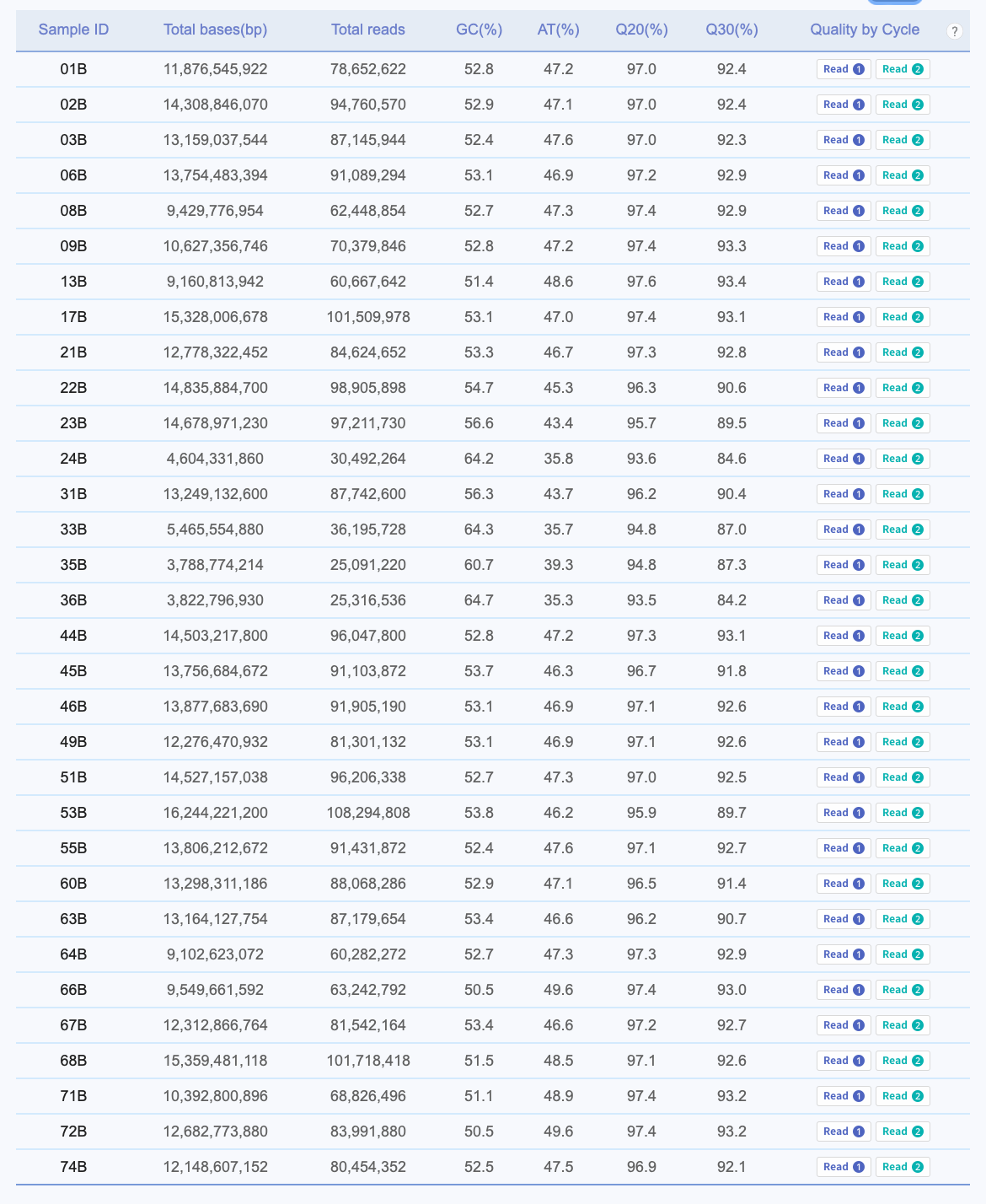
AN00025267 / AN00025268
AN00025267 / AN00025268 are paired RNA-seq / WGBS samples. Samples 1-36 are blood from cod held at 16C and samples 44-74 are blood from cod held at 0C. Note we also have lcWGS (~3x depth, a few samples higher) for the same cod individuals, should we want to, say, compare SNPs pulled from WGBS and lcWGS, or do a similar genetic + epigenetic variation paper (like the Silliman & Spencer Oly paper).
AN00025267
Link to AN00025267 report:
Link to data - https://owl.fish.washington.edu/nightingales/G_macrocephalus/AN00025267/
AN00025268
Link to Report
Link to data
Genome
https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_031168955.1/
Larger Effort
| Project | Type | Unique samples | FASTQ/FQ files | Naming schema | Examples | Status |
|---|---|---|---|---|---|---|
| 30-943133806 | RNA-seq | 80 | 160 | NN[_SUFFIX]_R{1,2}_001.fastq.gz, RESUB-###_R{1,2}_001.fastq.gz |
100_R1_001.fastq.gz, 19-G_R1_001.fastq.gz, RESUB-76_R1_001.fastq.gz |
Complete - WGCNA analysis identified temperature-responsive genes (0, 5, 9, 16C) |
| H202SC23041287 | lcWGS | 157 | 314* | GM##_*_{1,2}.fq.gz (sample GM##, multiple lanes/runs) |
GM1_CKDN230011839-1A_H5NNGDSX7_L1_1.fq.gz |
Complete - Population assignment, RDA, GWAS, eQTL analyses |
| 30-1149633765 | WGS | 350 | 700 | EXP###_R{1,2}_001.fastq.gz, MHW###_R{1,2}_001.fastq.gz |
EXP001_R1_001.fastq.gz, MHW001_R1_001.fastq.gz |
In progress - heatwave genetics study (2008-2023) + experimental resequencing |
| 30-1149634506 | WGS | 16 | 32 | ET##_R{1,2}_001.fastq.gz |
ET22_R1_001.fastq.gz |
In progress - pilot ecotype sequencing (shallow/deep) |
| AN00025267** | BS-seq | 32 | 64 | ##B_{1,2}.fastq.gz (paired reads as _1/_2) |
01B_1.fastq.gz, 01B_2.fastq.gz |
In progress - nf-core/methylseq pipeline (17.nextflow) |
| AN00025268** | BS-seq | 32 | 64 | ##B_{1,2}.fastq.gz (paired reads as _1/_2) |
01B_1.fastq.gz, 01B_2.fastq.gz |
Planned |
| 30-1067895835** | WGBS | 1 | 2 | SAMPLE_R{1,2}_001.fastq.gz |
1D11_R1_001.fastq.gz |
trial WGBS for Pacific cod methylation and depth, in progress |
Key Results to Date
- RNA-seq differential expression: Weighted gene correlation network analysis (WGCNA) identified genes expressed in the liver that are temperature-responsive, and those that correlate with juvenile cod performance (e.g. liver lipid content) within temperatures, particularly warming.
- Gene ontology enrichment: GO term enrichment analysis performed using DAVID on DEGs.
- Population genetics: lcWGS data analyzed to assign experimental fish to known Pacific cod spawning populations using WGSassign, PCA, which indicated that all were from the GOA spawning aggregation. Redundancy analysis (RDA) indicated that genetic variation associated with GOA populations (eGOA, wGOA) was not strongly linked to phenotypic responses in warming (16C), while there was some evidence for population-associated variation in cold acclimation (e.g. polar lipid concentrations). Additional genetic structure (within the wGOA population) existed that was not associated with the wGOA and eGOA populations that correlated with trait variation.
- Genome-Wide Association Study (GWAS): lcWGS data paired with per-individual traits (growth, condition, liver lipid content) measured across four temperatures (0-16C) identified genetic variants associated with trait variation.
- Growth and physiology: Significant treatment effects observed on growth rates (SGRww, SGRsl), condition indices (Kwet, HSI), and liver lipid composition. Temperature-dependent models were constructed.