This table identifies genes whose lncRNA-mediated regulation varies depending on miRNA expression. Note that current interaction metrics behave similarly or more strongly under randomized data and should be interpreted cautiously.
This table shows genes with the largest multi-regulator improvements. Current evidence suggests similar improvements can arise under randomized data; treat these results as hypothesis-generating rather than definitive.
These CSV files contain the detailed results of context-dependent regulatory analysis, including interaction statistics, p-values, context strengths, and regulatory relationships for each analyzed gene-regulator pair.
When empirical FDR is enabled, includes empirical_fdr_threshold, empirical_fdr_estimated, and empirical_fdr_significant.
How to assess confidence: prioritize interactions with empirical_fdr_significant == True (if available); otherwise, use context_strength as the primary effect-size metric, ideally cross-referenced against random-data null runs.
Correlation networks in the high-methylation context, defined using a sentinel CpG site.
Each row is a gene–regulator pair with a Pearson correlation (correlation) and raw p_value computed only within high-methylation samples.
How to assess confidence: these p-values are not corrected for multiple testing; treat them as exploratory. For higher confidence, focus on strong effect sizes (|correlation| close to 1) and/or overlap with FDR-supported context interactions from methylation_mirna_context.csv.
Correlation networks in the low-miRNA context, using a sentinel miRNA as the context-defining variable.
Each row is a gene–regulator pair with a Pearson correlation (correlation) and raw p_value computed only within low-miRNA samples.
How to assess confidence: as above, p-values are unadjusted across many tests; use them as a guide for ranking, not strict significance. Strong |correlation| and consistency with patterns seen in methylation_mirna_context.csv or across contexts provide more persuasive evidence.
lncrna_mirna_context.csv (if lncRNA context module is enabled):
Regression-based lncRNA–miRNA–gene context metrics analogous to methylation_mirna_context.csv.
Empirical comparisons with randomized data suggest that current interaction metrics here are more exploratory; interpret “context-dependent” calls cautiously.
How to assess confidence: use this table primarily for hypothesis generation or to find lncRNAs associated with genes that already have strong methylation–miRNA context signals.
multi_way_interactions.csv (if multi-way module is enabled):
Summarizes multi-regulator regression models for each gene, comparing a simple model to a full model with many regulators.
Contains improvement_from_regulators and an F-test interaction_p_value with a boolean has_significant_interactions.
How to assess confidence: current evidence shows that large improvements can also arise in randomized data; treat these results as exploratory and consider additional validation (e.g., overlap with simpler context metrics or external datasets).
Analysis Parameters
Parallel workers: 192
Data directory: /mmfs1/gscratch/scrubbed/sr320/github/ConTra/data/full-species-biomin/cleaned_ptua
This analysis successfully identified context-dependent regulatory interactions using optimized parallel processing. The results provide insights into how different regulatory layers interact in a context-specific manner.
Report generated by OptimizedContextDependentRegulationAnalysis on 2025-12-02 12:52:16
This table identifies genes whose lncRNA-mediated regulation varies depending on miRNA expression. Note that current interaction metrics behave similarly or more strongly under randomized data and should be interpreted cautiously.
Rank
Gene
lncRNA
miRNA
Improvement
Context Strength
1
(‘OG_18262’, ‘Peve_00044240’)
lncrna_lncRNA_10659
mirna_Cluster_6099
0.469
1.007
2
(‘OG_18262’, ‘Peve_00044240’)
lncrna_lncRNA_10659
mirna_Cluster_6918
0.458
0.989
3
(‘OG_13684’, ‘Peve_00018981’)
lncrna_lncRNA_12581
mirna_Cluster_6234
0.444
0.898
4
(‘OG_12320’, ‘Peve_00024570’)
lncrna_lncRNA_19767
mirna_Cluster_670
0.440
nan
5
(‘OG_12320’, ‘Peve_00024570’)
lncrna_lncRNA_19765
mirna_Cluster_670
0.439
nan
6
(‘OG_13526’, ‘Peve_00019641’)
lncrna_lncRNA_20807
mirna_Cluster_670
0.437
nan
7
(‘OG_13526’, ‘Peve_00019641’)
lncrna_lncRNA_23227
mirna_Cluster_670
0.437
nan
8
(‘OG_13684’, ‘Peve_00018981’)
lncrna_lncRNA_23131
mirna_Cluster_981
0.433
0.861
9
(‘OG_13684’, ‘Peve_00018981’)
lncrna_lncRNA_12581
mirna_Cluster_5257
0.433
0.975
10
(‘OG_13684’, ‘Peve_00018981’)
lncrna_lncRNA_12581
mirna_Cluster_6235
0.432
0.987
Multi-Way Interaction Analysis (Exploratory)
Total genes analyzed: 553
Genes with significant interactions (F-test): 0
Mean improvement from interactions: 0.574
Top 10 Multi-Way Interactions (Exploratory)
This table shows genes with the largest multi-regulator improvements. Current evidence suggests similar improvements can arise under randomized data; treat these results as hypothesis-generating rather than definitive.
These CSV files contain the detailed results of context-dependent regulatory analysis, including interaction statistics, p-values, context strengths, and regulatory relationships for each analyzed gene-regulator pair.
When empirical FDR is enabled, includes empirical_fdr_threshold, empirical_fdr_estimated, and empirical_fdr_significant.
How to assess confidence: prioritize interactions with empirical_fdr_significant == True (if available); otherwise, use context_strength as the primary effect-size metric, ideally cross-referenced against random-data null runs.
Correlation networks in the high-methylation context, defined using a sentinel CpG site.
Each row is a gene–regulator pair with a Pearson correlation (correlation) and raw p_value computed only within high-methylation samples.
How to assess confidence: these p-values are not corrected for multiple testing; treat them as exploratory. For higher confidence, focus on strong effect sizes (|correlation| close to 1) and/or overlap with FDR-supported context interactions from methylation_mirna_context.csv.
Correlation networks in the low-miRNA context, using a sentinel miRNA as the context-defining variable.
Each row is a gene–regulator pair with a Pearson correlation (correlation) and raw p_value computed only within low-miRNA samples.
How to assess confidence: as above, p-values are unadjusted across many tests; use them as a guide for ranking, not strict significance. Strong |correlation| and consistency with patterns seen in methylation_mirna_context.csv or across contexts provide more persuasive evidence.
lncrna_mirna_context.csv (if lncRNA context module is enabled):
Regression-based lncRNA–miRNA–gene context metrics analogous to methylation_mirna_context.csv.
Empirical comparisons with randomized data suggest that current interaction metrics here are more exploratory; interpret “context-dependent” calls cautiously.
How to assess confidence: use this table primarily for hypothesis generation or to find lncRNAs associated with genes that already have strong methylation–miRNA context signals.
multi_way_interactions.csv (if multi-way module is enabled):
Summarizes multi-regulator regression models for each gene, comparing a simple model to a full model with many regulators.
Contains improvement_from_regulators and an F-test interaction_p_value with a boolean has_significant_interactions.
How to assess confidence: current evidence shows that large improvements can also arise in randomized data; treat these results as exploratory and consider additional validation (e.g., overlap with simpler context metrics or external datasets).
Analysis Parameters
Parallel workers: 192
Data directory: /mmfs1/gscratch/scrubbed/sr320/github/ConTra/data/full-species-biomin/cleaned_peve
This analysis successfully identified context-dependent regulatory interactions using optimized parallel processing. The results provide insights into how different regulatory layers interact in a context-specific manner.
Report generated by OptimizedContextDependentRegulationAnalysis on 2025-11-30 19:41:04
This report presents the results of context-dependent regulatory interaction analysis between: - Gene expression - miRNA expression - lncRNA expression - DNA methylation
Analysis Overview
Parallel Processing: 192 CPU cores
Available RAM: 2940.2 GB
Datasets Loaded: 4 data types
Gene: 39 samples × 517 features
Lncrna: 39 samples × 15559 features
Mirna: 39 samples × 51 features
Methylation: 39 samples × 66428 features
Results Summary
Methylation-miRNA Context Analysis
Total interactions analyzed: 51150
Context-dependent interactions (F-test): 8765
Mean improvement from interaction: 0.027
Mean context strength: 0.346
Top 10 Methylation-miRNA Interactions (by Context Strength)
This table shows genes whose regulation by DNA methylation is context-dependent on miRNA expression levels, ranked by context strength.
Rank
Gene
Methylation
miRNA
Improvement
Context Strength
Empirical FDR Sig.
1
(‘OG_05303’, ‘FUN_023974’)
methylation_CpG_ptg000001l_9710125
mirna_Cluster_3109
0.060
1.881
False
2
(‘OG_07265’, ‘FUN_033056’)
methylation_CpG_ptg000021l_10139814
mirna_Cluster_3109
0.018
1.875
False
3
(‘OG_14547’, ‘FUN_019026’)
methylation_CpG_ptg000023l_11736283
mirna_Cluster_3109
0.023
1.875
False
4
(‘OG_07265’, ‘FUN_033056’)
methylation_CpG_ptg000018l_7377398
mirna_Cluster_3109
0.067
1.854
False
5
(‘OG_10509’, ‘FUN_002531’)
methylation_CpG_ptg000031l_5310815
mirna_Cluster_3109
0.004
1.837
False
6
(‘OG_06610’, ‘FUN_029760’)
methylation_CpG_ptg000031l_5310762
mirna_Cluster_3109
0.004
1.833
False
7
(‘OG_10811’, ‘FUN_006888’)
methylation_CpG_ptg000023l_8252944
mirna_Cluster_3109
0.006
1.756
False
8
(‘OG_14711’, ‘FUN_024326’)
methylation_CpG_ptg000012l_17885415
mirna_Cluster_3109
0.045
1.672
False
9
(‘OG_10811’, ‘FUN_006888’)
methylation_CpG_ptg000027l_7575122
mirna_Cluster_3109
0.015
1.660
False
10
(‘OG_06610’, ‘FUN_029760’)
methylation_CpG_ptg000021l_6263529
mirna_Cluster_3109
0.012
1.657
False
lncRNA-miRNA Context Analysis (Exploratory)
Total interactions analyzed: 51150
Context-dependent interactions (F-test): 8365
Mean improvement from interaction: 0.021
Mean context strength: 0.270
Top 10 lncRNA-miRNA Interactions (Exploratory)
This table identifies genes whose lncRNA-mediated regulation varies depending on miRNA expression. Note that current interaction metrics behave similarly or more strongly under randomized data and should be interpreted cautiously.
Rank
Gene
lncRNA
miRNA
Improvement
Context Strength
1
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_48837
mirna_Cluster_14165
0.420
1.162
2
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_48837
mirna_Cluster_14146
0.407
1.082
3
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_48837
mirna_Cluster_3226
0.406
0.969
4
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_48837
mirna_Cluster_10452
0.400
0.910
5
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_23415
mirna_Cluster_9512
0.394
1.033
6
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_23414
mirna_Cluster_9512
0.392
1.021
7
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_23416
mirna_Cluster_9512
0.391
1.028
8
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_54028
mirna_Cluster_4752
0.387
0.966
9
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_48837
mirna_Cluster_4752
0.379
1.288
10
(‘OG_10579’, ‘FUN_003891’)
lncrna_lncRNA_44695
mirna_Cluster_4752
0.377
0.834
Multi-Way Interaction Analysis (Exploratory)
Total genes analyzed: 517
Genes with significant interactions (F-test): 0
Mean improvement from interactions: 0.750
Top 10 Multi-Way Interactions (Exploratory)
This table shows genes with the largest multi-regulator improvements. Current evidence suggests similar improvements can arise under randomized data; treat these results as hypothesis-generating rather than definitive.
These CSV files contain the detailed results of context-dependent regulatory analysis, including interaction statistics, p-values, context strengths, and regulatory relationships for each analyzed gene-regulator pair.
When empirical FDR is enabled, includes empirical_fdr_threshold, empirical_fdr_estimated, and empirical_fdr_significant.
How to assess confidence: prioritize interactions with empirical_fdr_significant == True (if available); otherwise, use context_strength as the primary effect-size metric, ideally cross-referenced against random-data null runs.
Correlation networks in the high-methylation context, defined using a sentinel CpG site.
Each row is a gene–regulator pair with a Pearson correlation (correlation) and raw p_value computed only within high-methylation samples.
How to assess confidence: these p-values are not corrected for multiple testing; treat them as exploratory. For higher confidence, focus on strong effect sizes (|correlation| close to 1) and/or overlap with FDR-supported context interactions from methylation_mirna_context.csv.
Correlation networks in the low-miRNA context, using a sentinel miRNA as the context-defining variable.
Each row is a gene–regulator pair with a Pearson correlation (correlation) and raw p_value computed only within low-miRNA samples.
How to assess confidence: as above, p-values are unadjusted across many tests; use them as a guide for ranking, not strict significance. Strong |correlation| and consistency with patterns seen in methylation_mirna_context.csv or across contexts provide more persuasive evidence.
lncrna_mirna_context.csv (if lncRNA context module is enabled):
Regression-based lncRNA–miRNA–gene context metrics analogous to methylation_mirna_context.csv.
Empirical comparisons with randomized data suggest that current interaction metrics here are more exploratory; interpret “context-dependent” calls cautiously.
How to assess confidence: use this table primarily for hypothesis generation or to find lncRNAs associated with genes that already have strong methylation–miRNA context signals.
multi_way_interactions.csv (if multi-way module is enabled):
Summarizes multi-regulator regression models for each gene, comparing a simple model to a full model with many regulators.
Contains improvement_from_regulators and an F-test interaction_p_value with a boolean has_significant_interactions.
How to assess confidence: current evidence shows that large improvements can also arise in randomized data; treat these results as exploratory and consider additional validation (e.g., overlap with simpler context metrics or external datasets).
Analysis Parameters
Parallel workers: 192
Data directory: /mmfs1/gscratch/scrubbed/sr320/github/ConTra/data/full-species-biomin/cleaned_apul
This analysis successfully identified context-dependent regulatory interactions using optimized parallel processing. The results provide insights into how different regulatory layers interact in a context-specific manner.
Report generated by OptimizedContextDependentRegulationAnalysis on 2025-12-01 11:13:07
Source Code
---title: "Calcifying Contra"description: "and beyond"categories: [e5, coral]#citation: date: 11-29-2025image: http://gannet.fish.washington.edu/seashell/snaps/Monosnap_Image_2025-11-30_08-27-36.png # finding a good imageauthor: - name: Steven Roberts url: orcid: 0000-0001-8302-1138 affiliation: Professor, UW - School of Aquatic and Fishery Sciences affiliation-url: https://robertslab.info #url: # self-defineddraft: false # setting this to `true` will prevent your post from appearing on your listing page until you're ready!format: html: toc: true # ← enable TOC toc-location: left # or: right, body toc-depth: 3 # how many heading levels to show code-fold: FALSE code-tools: true code-copy: true highlight-style: github code-overflow: wrap#runtime: shiny---First off in time-series-calcificationhttps://github.com/urol-e5/timeseries-molecular-calcification/blob/main/M-multi-species/scripts/34-biomin-pathway-compare.rmd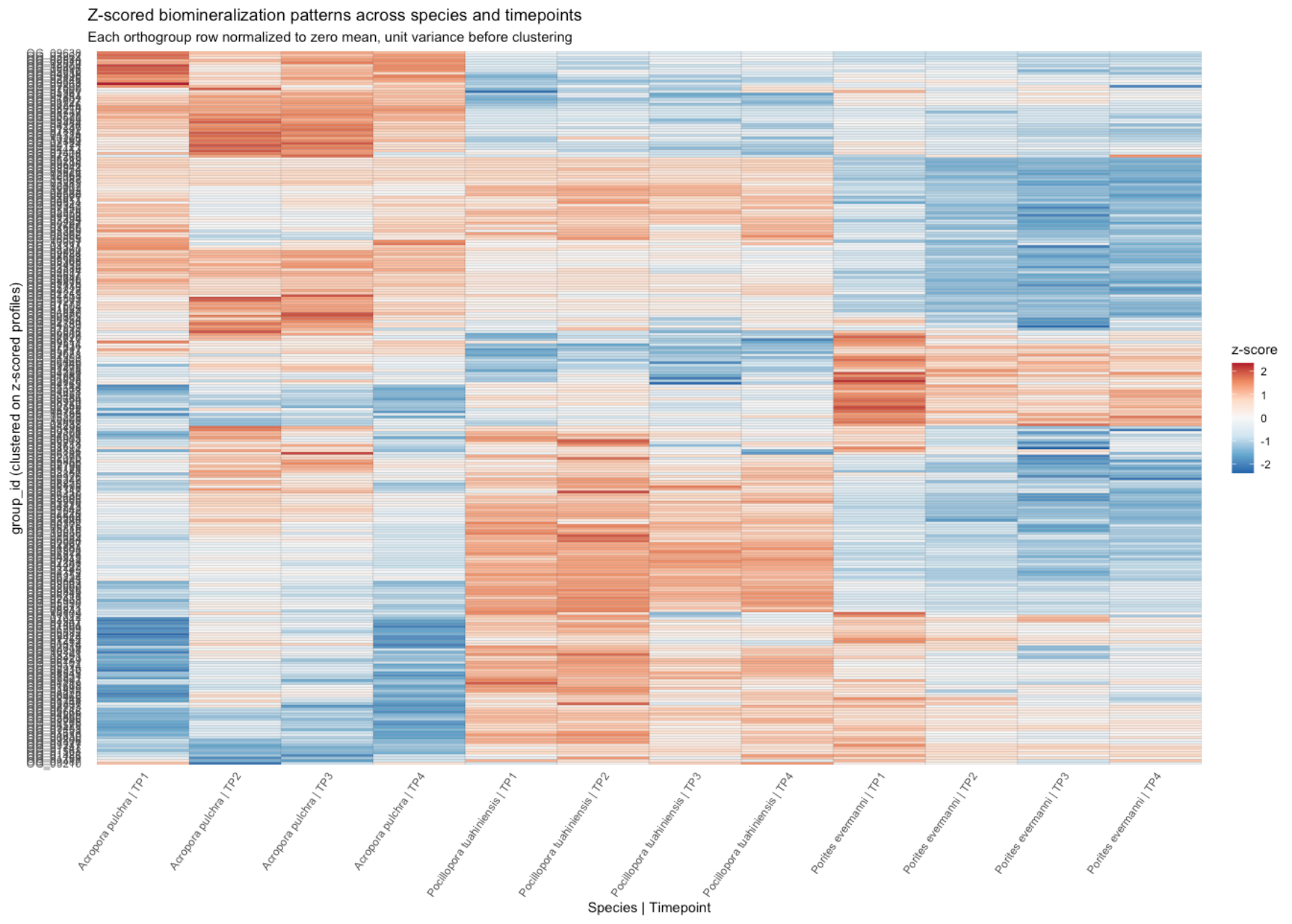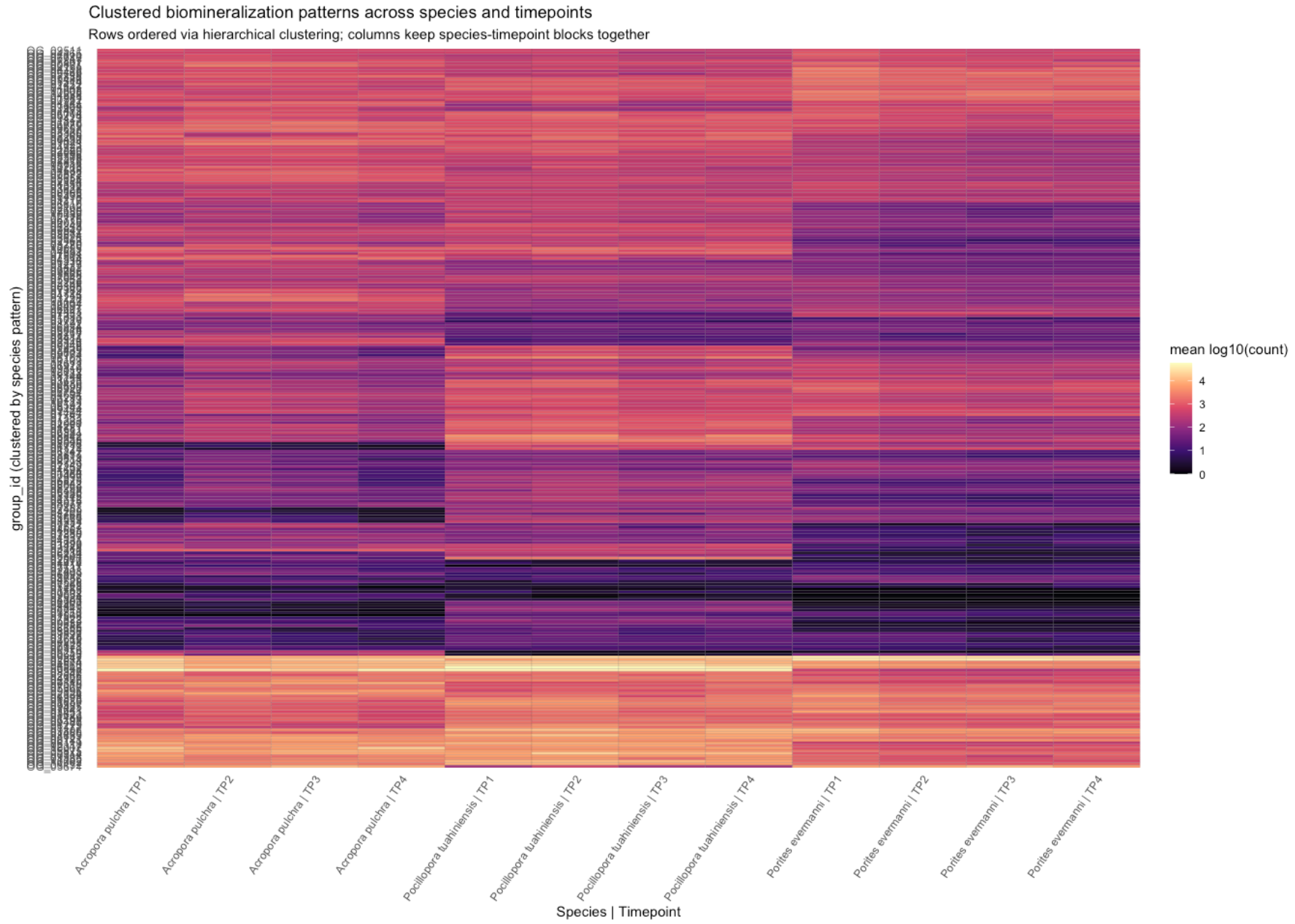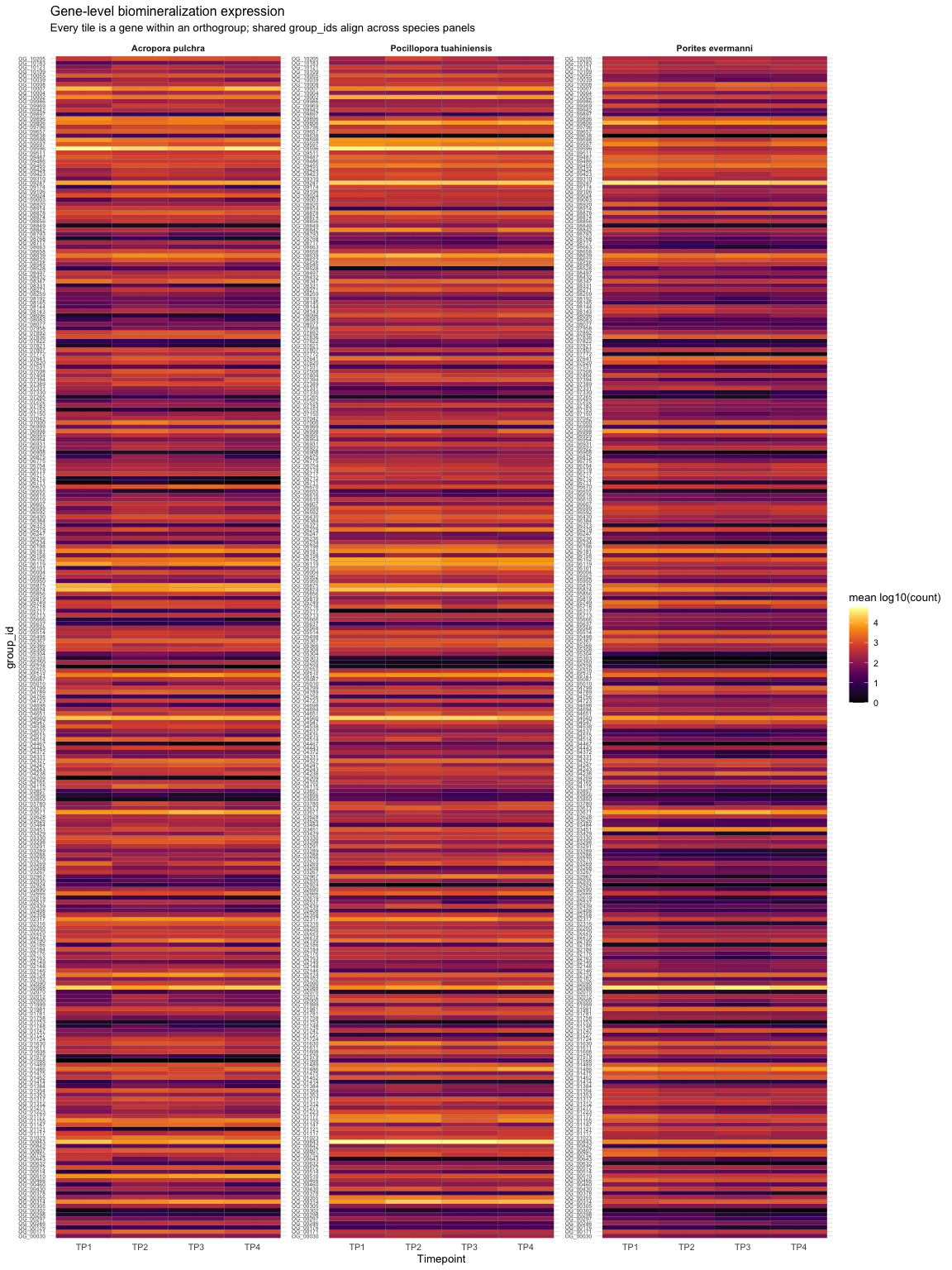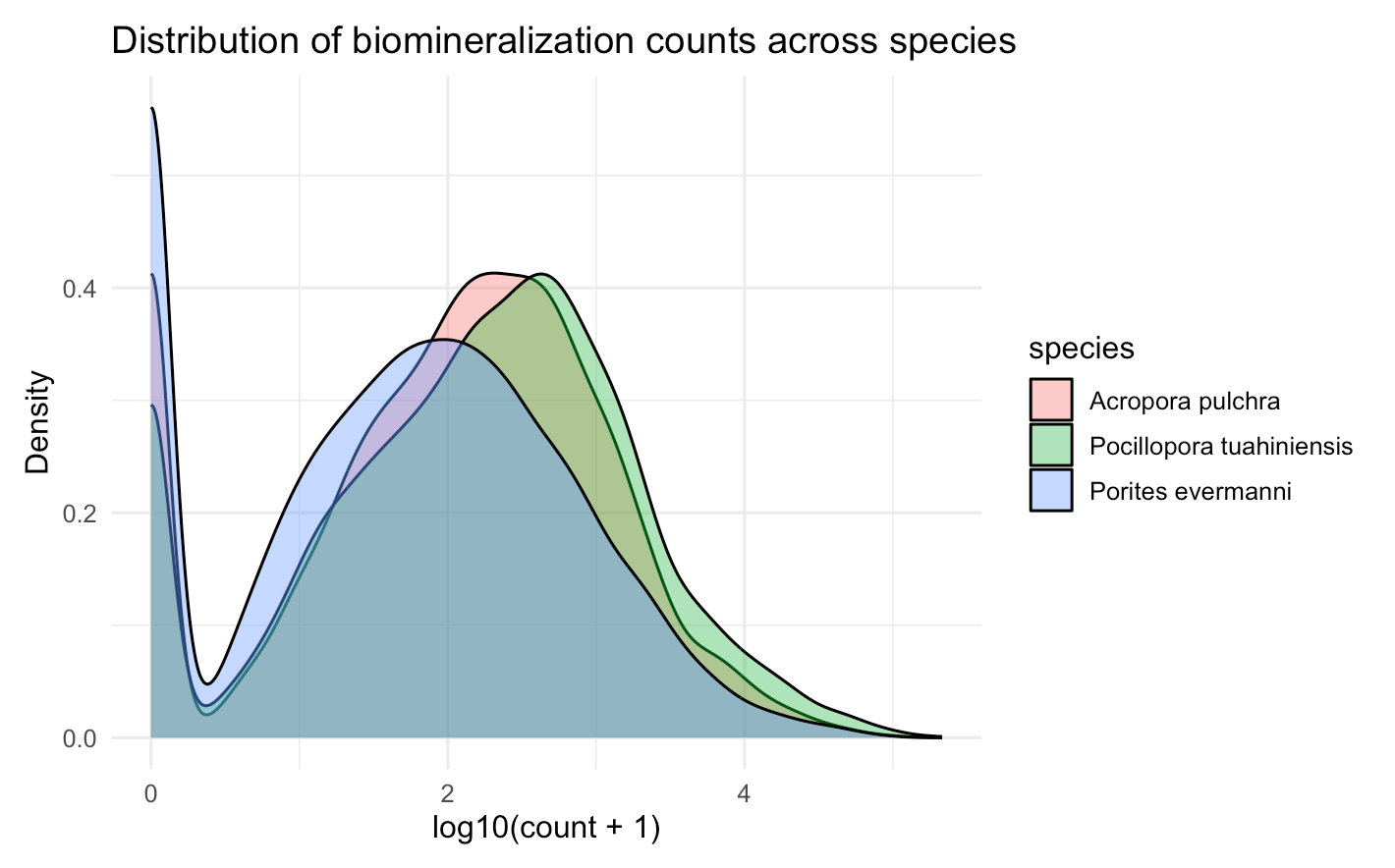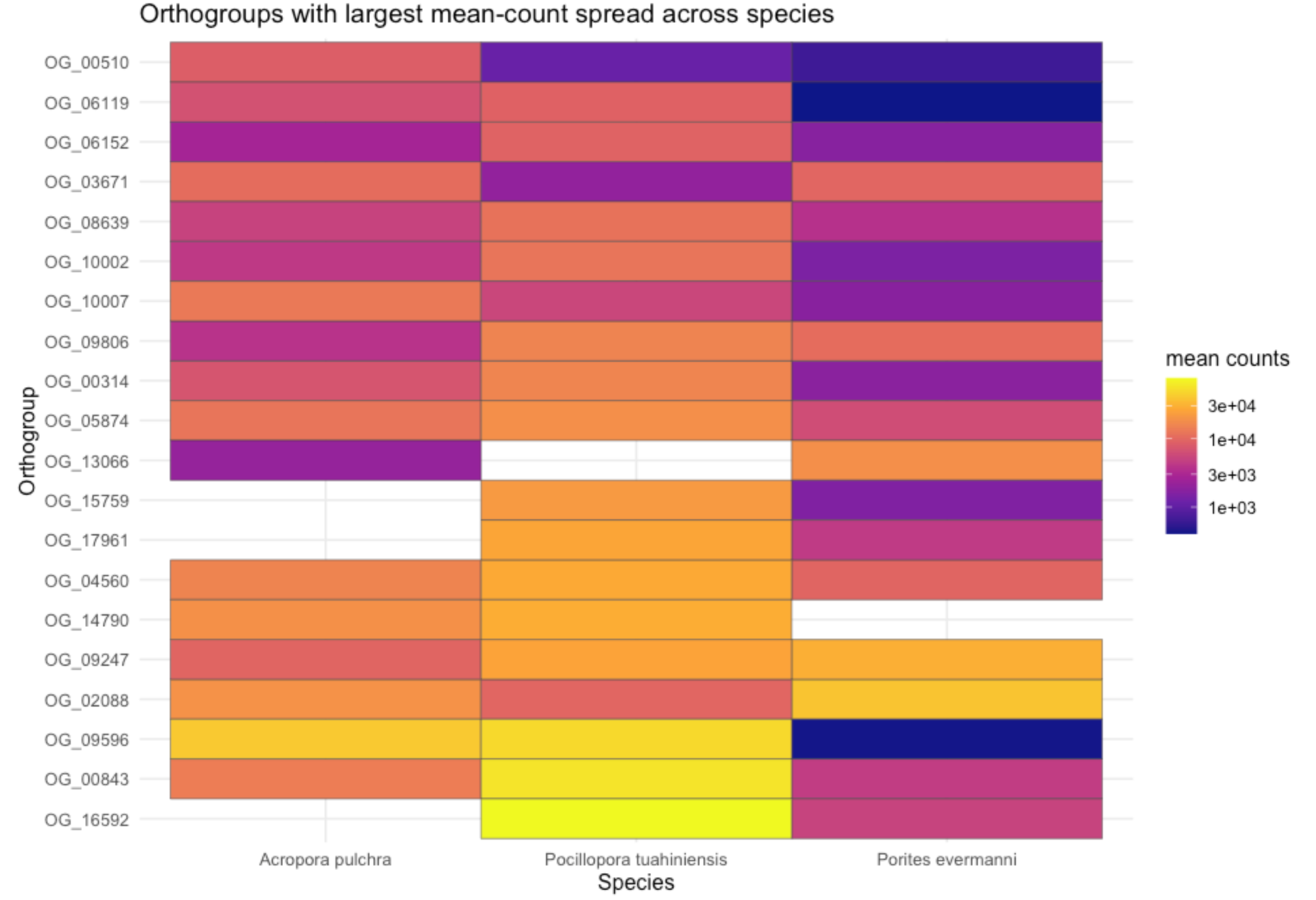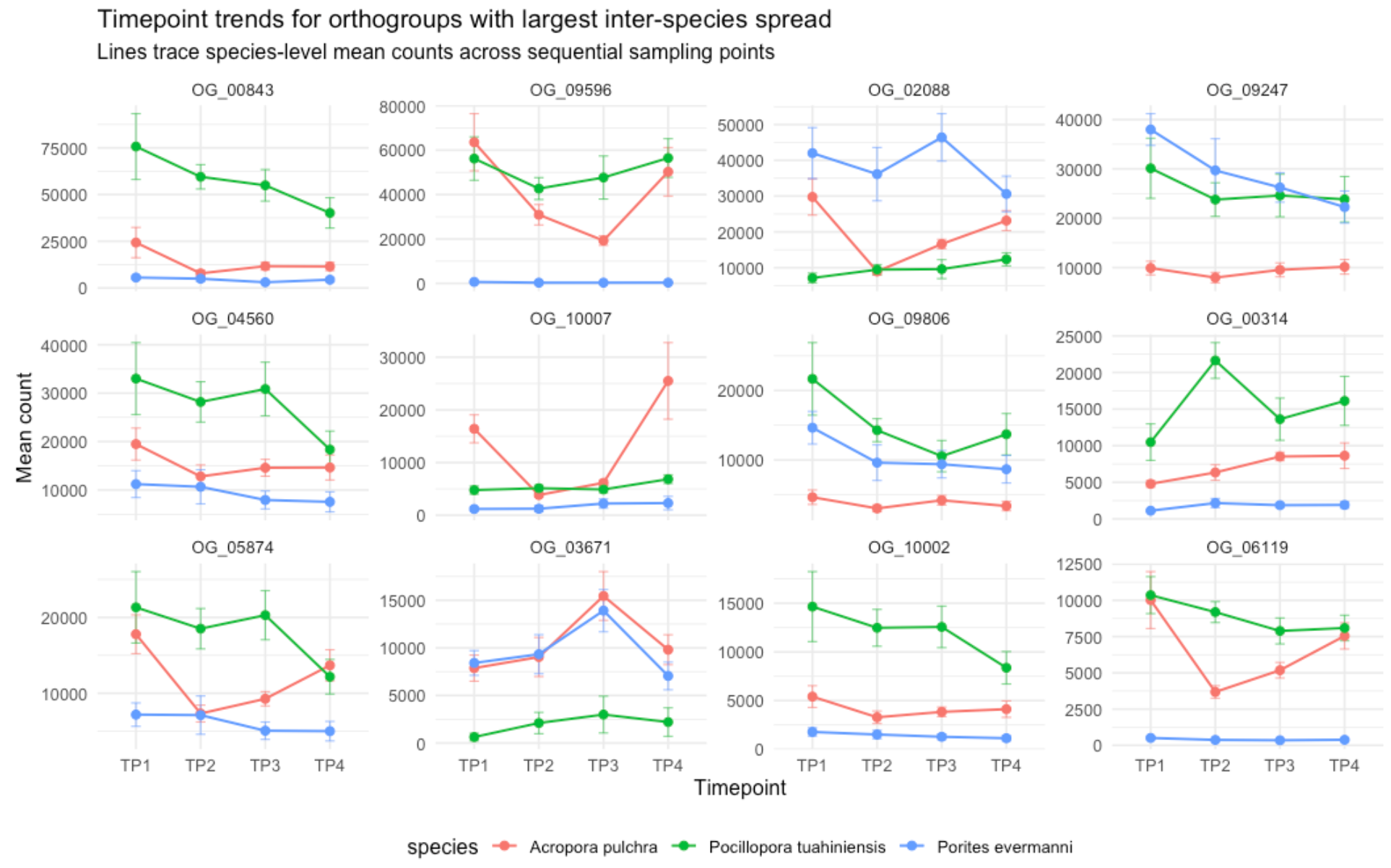# ContraThen took onto ConTra... with new script``` Duro:ConTra sr320$ python3 code/run_biomin_species_comparison.py ======================================================================🧬 CROSS-SPECIES BIOMIN CONTEXT-DEPENDENT ANALYSIS======================================================================Workspace: /Users/sr320/GitHub/ConTraComparison output: /Users/sr320/GitHub/ConTra/output/biomin_comparison_20251129_144655======================================================================`````` Using context_dependent_analysis_20251129_192544 for apul Using context_dependent_analysis_20251129_162258 for peve Using context_dependent_analysis_20251129_144655 for ptua============================================================LOADING SPECIES RESULTS============================================================📂 Loading A. pulchra... ✅ Loaded methylation_mirna_context.csv: 33270 rows📂 Loading P. evermanni... ✅ Loaded methylation_mirna_context.csv: 27150 rows📂 Loading P. tuahiniensis... ✅ Loaded methylation_mirna_context.csv: 51150 rows============================================================COMPARING METHYLATION-MIRNA CONTEXT ACROSS SPECIES============================================================📊 Summary: Total unique OGs across all species: 611 OGs in 2+ species: 358 OGs in all 3 species: 100✅ Saved comparison table: /Users/sr320/GitHub/ConTra/output/biomin_comparison_20251130_052506/cross_species_methylation_mirna_comparison.csv🏆 Top 15 Conserved Context-Dependent OGs (in 2+ species): og_id n_species mean_context_strength apul_context_strength peve_context_strength ptua_context_strengthOG_01155 3 1.310726 1.521005 0.996267 1.414906OG_02619 3 1.295301 1.468612 1.024492 1.392800OG_06119 3 1.236816 0.971755 1.252635 1.486059OG_08793 3 1.236346 0.808764 1.450764 1.449511OG_05366 3 1.199557 0.939276 1.174450 1.484946OG_03291 3 1.199266 1.186752 0.799569 1.611478OG_05637 3 1.192262 1.166134 1.165108 1.245543OG_01452 3 1.191146 1.526632 0.922735 1.124072OG_07153 3 1.174770 0.979025 1.358085 1.187201OG_01414 3 1.161650 1.680689 0.775683 1.028577OG_01753 3 1.160818 1.140792 1.097454 1.244207OG_08663 3 1.138407 0.935220 1.247335 1.232668OG_09796 3 1.133137 1.016265 1.074893 1.308253OG_04467 3 1.121429 1.075908 1.254588 1.033790OG_07892 3 1.113713 1.090661 0.877698 1.372780============================================================GENERATING VISUALIZATIONS============================================================/Users/sr320/GitHub/ConTra/code/run_biomin_species_comparison.py:397: FutureWarning: Passing `palette` without assigning `hue` is deprecated and will be removed in v0.14.0. Assign the `x` variable to `hue` and set `legend=False` for the same effect. sns.violinplot(data=strength_df, x='species', y='context_strength',✅ Saved comparison plots: /Users/sr320/GitHub/ConTra/output/biomin_comparison_20251130_052506/plots/species_comparison_overview.png✅ Saved comparison report: /Users/sr320/GitHub/ConTra/output/biomin_comparison_20251130_052506/cross_species_comparison_report.md======================================================================🎉 CROSS-SPECIES COMPARISON COMPLETE!======================================================================Results saved to: /Users/sr320/GitHub/ConTra/output/biomin_comparison_20251130_052506======================================================================```## **What the script does:**1. **Runs analysis on all 3 species** in [full-species-biomin](vscode-file://vscode-app/Applications/Visual%20Studio%20Code.app/Contents/Resources/app/out/vs/code/electron-browser/workbench/workbench.html): - A. pulchra (apul) - P. evermanni (peve) - P. tuahiniensis (ptua)2. **Extracts OG IDs** from the results using regex to parse formats like: - `"('OG_13910', 'FUN_002435')"` - `OG_13910`3. **Compares results across species**: - Finds OGs present in 2+ species - Ranks by conservation and context strength - Calculates correlations between species4. **Generates outputs**: - `cross_species_methylation_mirna_comparison.csv` - Full comparison table - `plots/species_comparison_overview.png` - 4-panel visualization - `cross_species_comparison_report.md` - Summary report------------------------------------------------------------------------# Cross-Species Biomin Context-Dependent Analysis Comparison**Generated:** 2025-11-30 05:25:07## OverviewThis report compares context-dependent regulatory interactions across three coral species:- **A. pulchra** (`apul`)- **P. evermanni** (`peve`)- **P. tuahiniensis** (`ptua`)## Per-Species Summary### A. pulchra (apul)- Total methylation-miRNA interactions: 33270- Context-dependent interactions: 3039- Mean context strength: 0.311- Unique OGs: 334### P. evermanni (peve)- Total methylation-miRNA interactions: 27150- Context-dependent interactions: 3935- Mean context strength: 0.371- Unique OGs: 309### P. tuahiniensis (ptua)- Total methylation-miRNA interactions: 51150- Context-dependent interactions: 8765- Mean context strength: 0.346- Unique OGs: 474## Cross-Species Conservation- **Total unique OGs:** 611- **OGs in 2+ species:** 358 (58.6%)- **OGs in all 3 species:** 100 (16.4%)## InterpretationOGs (orthologous groups) that show context-dependent regulation in multiple species suggest:1. **Conserved regulatory mechanisms** - These genes may have fundamental roles where regulation is evolutionarily maintained2. **Robust biological signals** - Conservation across species reduces the likelihood of false positives3. **Candidates for functional validation** - These genes are high-priority targets for experimental follow-up## Files Generated- `cross_species_methylation_mirna_comparison.csv` - Full comparison table- `plots/species_comparison_overview.png` - Visualization of cross-species patterns- `cross_species_comparison_report.md` - This report------------------------------------------------------------------------```{r}library(tidyverse)library(reactable)library(htmltools)url <-"https://gannet.fish.washington.edu/v1_web/owlshell/bu-github/ConTra/output/biomin_comparison_20251130_052506/cross_species_methylation_mirna_comparison.csv"df <-read_csv(url, show_col_types =FALSE)# Format all numeric columns to 2 digits after decimaldf_fmt <- df %>%mutate(across(where(is.numeric), ~sprintf("%.2f", .x)))reactable( df_fmt,searchable =TRUE,filterable =TRUE,pagination =TRUE,highlight =TRUE,striped =TRUE,defaultPageSize =20,sortable =TRUE,theme = reactable::reactableTheme(highlightColor ="#EAF2F8" ))```# Ptua Context-Dependent Regulation Analysis Report**Generated:** 2025-12-02 12:52:16**Analysis ID:** 20251201_160901## Executive SummaryThis report presents the results of context-dependent regulatory interaction analysis between:- Gene expression- miRNA expression- lncRNA expression- DNA methylation## Analysis Overview- **Parallel Processing:** 192 CPU cores- **Available RAM:** 2941.9 GB- **Datasets Loaded:** 4 data types - Gene: 32 samples × 483 features - Lncrna: 32 samples × 11236 features - Mirna: 32 samples × 40 features - Methylation: 32 samples × 263324 features## Results Summary### Methylation-miRNA Context Analysis- **Total interactions analyzed:** 33270- **Context-dependent interactions (F-test):** 3039- **Mean improvement from interaction:** 0.017- **Mean context strength:** 0.311#### Top 10 Methylation-miRNA Interactions (by Context Strength)*This table shows genes whose regulation by DNA methylation is context-dependent on miRNA expression levels, ranked by context strength.*| Rank | Gene | Methylation | miRNA | Improvement | Context Strength | Empirical FDR Sig. ||------|------|-------------|-------|-------------|------------------|--------------------|| 1 | ('OG_02537', 'Pocillopora_meandrina_HIv1___TS.g26115.t1') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000010_2525456 | mirna_Cluster_4609 | 0.043 | 1.805 | False || 2 | ('OG_08920', 'Pocillopora_meandrina_HIv1___RNAseq.g1090.t1') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000017_5066475 | mirna_Cluster_4094 | 0.016 | 1.770 | False || 3 | ('OG_01414', 'Pocillopora_meandrina_HIv1___TS.g28923.t1a') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000003_1369854 | mirna_Cluster_4826 | 0.015 | 1.681 | False || 4 | ('OG_04723', 'Pocillopora_meandrina_HIv1___RNAseq.g4434.t2') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000000_18894187 | mirna_Cluster_4826 | 0.131 | 1.600 | False || 5 | ('OG_15303', 'Pocillopora_meandrina_HIv1___TS.g5438.t1') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000018_8279715 | mirna_Cluster_6807 | 0.004 | 1.583 | False || 6 | ('OG_14804', 'Pocillopora_meandrina_HIv1___RNAseq.g24442.t1') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000006_12397256 | mirna_Cluster_4485 | 0.005 | 1.562 | False || 7 | ('OG_13910', 'Pocillopora_meandrina_HIv1___RNAseq.g778.t1') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000010_5065428 | mirna_Cluster_3415 | 0.002 | 1.561 | False || 8 | ('OG_08920', 'Pocillopora_meandrina_HIv1___RNAseq.g1090.t1') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000017_5066475 | mirna_Cluster_5123 | 0.040 | 1.545 | False || 9 | ('OG_01452', 'Pocillopora_meandrina_HIv1___RNAseq.g19573.t1') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000000_10732016 | mirna_Cluster_4826 | 0.265 | 1.527 | False || 10 | ('OG_01155', 'Pocillopora_meandrina_HIv1___RNAseq.g6698.t2') | methylation_CpG_Pocillopora_meandrina_HIv1___Sc0000011_3176997 | mirna_Cluster_4485 | 0.045 | 1.521 | False |### lncRNA-miRNA Context Analysis (Exploratory)- **Total interactions analyzed:** 33270- **Context-dependent interactions (F-test):** 4724- **Mean improvement from interaction:** 0.018- **Mean context strength:** 0.229#### Top 10 lncRNA-miRNA Interactions (Exploratory)*This table identifies genes whose lncRNA-mediated regulation varies depending on miRNA expression. Note that current interaction metrics behave similarly or more strongly under randomized data and should be interpreted cautiously.*| Rank | Gene | lncRNA | miRNA | Improvement | Context Strength ||------|------|--------|-------|-------------|------------------|| 1 | ('OG_14763', 'Pocillopora_meandrina_HIv1___RNAseq.g13186.t1') | lncrna_lncRNA_12271 | mirna_Cluster_6456 | 0.376 | nan || 2 | ('OG_14681', 'Pocillopora_meandrina_HIv1___TS.g17155.t1') | lncrna_lncRNA_30484 | mirna_Cluster_1159 | 0.360 | nan || 3 | ('OG_14763', 'Pocillopora_meandrina_HIv1___RNAseq.g13186.t1') | lncrna_lncRNA_12271 | mirna_Cluster_4 | 0.356 | nan || 4 | ('OG_14681', 'Pocillopora_meandrina_HIv1___TS.g17155.t1') | lncrna_lncRNA_5394 | mirna_Cluster_1159 | 0.354 | nan || 5 | ('OG_14681', 'Pocillopora_meandrina_HIv1___TS.g17155.t1') | lncrna_lncRNA_5394 | mirna_Cluster_3713 | 0.340 | nan || 6 | ('OG_14763', 'Pocillopora_meandrina_HIv1___RNAseq.g13186.t1') | lncrna_lncRNA_29138 | mirna_Cluster_6456 | 0.334 | nan || 7 | ('OG_14681', 'Pocillopora_meandrina_HIv1___TS.g17155.t1') | lncrna_lncRNA_30480 | mirna_Cluster_3713 | 0.333 | nan || 8 | ('OG_14763', 'Pocillopora_meandrina_HIv1___RNAseq.g13186.t1') | lncrna_lncRNA_29138 | mirna_Cluster_4 | 0.332 | nan || 9 | ('OG_14681', 'Pocillopora_meandrina_HIv1___TS.g17155.t1') | lncrna_lncRNA_30481 | mirna_Cluster_3713 | 0.329 | nan || 10 | ('OG_14763', 'Pocillopora_meandrina_HIv1___RNAseq.g13186.t1') | lncrna_lncRNA_29132 | mirna_Cluster_6456 | 0.326 | nan |### Multi-Way Interaction Analysis (Exploratory)- **Total genes analyzed:** 483- **Genes with significant interactions (F-test):** 0- **Mean improvement from interactions:** 0.597#### Top 10 Multi-Way Interactions (Exploratory)*This table shows genes with the largest multi-regulator improvements. Current evidence suggests similar improvements can arise under randomized data; treat these results as hypothesis-generating rather than definitive.*| Rank | Gene | Improvement | Significant Interactions ||------|------|-------------|------------------------|| 1 | ('OG_07000', 'Pocillopora_meandrina_HIv1___RNAseq.g5328.t1') | 0.912 | False || 2 | ('OG_02317', 'Pocillopora_meandrina_HIv1___TS.g30016.t1') | 0.912 | False || 3 | ('OG_08545', 'Pocillopora_meandrina_HIv1___RNAseq.g1314.t1') | 0.912 | False || 4 | ('OG_00466', 'Pocillopora_meandrina_HIv1___TS.g4618.t1') | 0.911 | False || 5 | ('OG_07822', 'Pocillopora_meandrina_HIv1___RNAseq.g27709.t1') | 0.910 | False || 6 | ('OG_04209', 'Pocillopora_meandrina_HIv1___RNAseq.g10045.t1') | 0.908 | False || 7 | ('OG_01117', 'Pocillopora_meandrina_HIv1___TS.g20470.t1') | 0.907 | False || 8 | ('OG_17014', 'Pocillopora_meandrina_HIv1___RNAseq.g15484.t1') | 0.907 | False || 9 | ('OG_02666', 'Pocillopora_meandrina_HIv1___RNAseq.g21004.t1') | 0.906 | False || 10 | ('OG_07042', 'Pocillopora_meandrina_HIv1___TS.g12785.t1') | 0.906 | False |## Data Files<https://gannet.fish.washington.edu/v1_web/owlshell/bu-github/ConTra/output/context_dependent_analysis_20251201_160901/tables/>The following data files were generated:These CSV files contain the detailed results of context-dependent regulatory analysis, including interaction statistics, p-values, context strengths, and regulatory relationships for each analyzed gene-regulator pair.- **high_methylation_gene_lncrna_correlations.csv** (130713.8 KB)- **high_methylation_gene_methylation_correlations.csv** (1814558.8 KB)- **high_methylation_gene_mirna_correlations.csv** (394.4 KB)- **high_mirna_gene_lncrna_correlations.csv** (88440.6 KB)- **high_mirna_gene_methylation_correlations.csv** (1713259.1 KB)- **high_mirna_gene_mirna_correlations.csv** (377.4 KB)- **lncrna_mirna_context.csv** (9879.4 KB)- **low_mirna_gene_lncrna_correlations.csv** (187585.6 KB)- **low_mirna_gene_methylation_correlations.csv** (1766258.4 KB)- **low_mirna_gene_mirna_correlations.csv** (966.3 KB)- **methylation_mirna_context.csv** (11173.1 KB)- **multi_way_interactions.csv** (1218.8 KB)### Table Definitions and Statistical ConfidenceBelow is a brief description of the main table types and how to assess statistical confidence for each:- **methylation_mirna_context.csv**: - One row per gene–CpG–miRNA triplet. - Contains regression-based metrics (`r2_*`, `improvement_from_regulator2`, `improvement_from_interaction`), conditional correlations (`corr_high_regulator2`, `corr_low_regulator2`), and `context_strength`. - When empirical FDR is enabled, includes `empirical_fdr_threshold`, `empirical_fdr_estimated`, and `empirical_fdr_significant`. - **How to assess confidence**: prioritize interactions with `empirical_fdr_significant == True` (if available); otherwise, use `context_strength` as the primary effect-size metric, ideally cross-referenced against random-data null runs.- **high_methylation_gene_methylation_correlations.csv** / **high_methylation_gene_mirna_correlations.csv** / **high_methylation_gene_lncrna_correlations.csv**: - Correlation networks in the **high-methylation context**, defined using a sentinel CpG site. - Each row is a gene–regulator pair with a Pearson correlation (`correlation`) and raw `p_value` computed only within high-methylation samples. - **How to assess confidence**: these p-values are not corrected for multiple testing; treat them as exploratory. For higher confidence, focus on strong effect sizes (|correlation| close to 1) and/or overlap with FDR-supported context interactions from `methylation_mirna_context.csv`.- **low_mirna_gene_methylation_correlations.csv** / **low_mirna_gene_mirna_correlations.csv** / **low_mirna_gene_lncrna_correlations.csv**: - Correlation networks in the **low-miRNA context**, using a sentinel miRNA as the context-defining variable. - Each row is a gene–regulator pair with a Pearson correlation (`correlation`) and raw `p_value` computed only within low-miRNA samples. - **How to assess confidence**: as above, p-values are unadjusted across many tests; use them as a guide for ranking, not strict significance. Strong |correlation| and consistency with patterns seen in `methylation_mirna_context.csv` or across contexts provide more persuasive evidence.- **lncrna_mirna_context.csv** (if lncRNA context module is enabled): - Regression-based lncRNA–miRNA–gene context metrics analogous to `methylation_mirna_context.csv`. - Empirical comparisons with randomized data suggest that current interaction metrics here are more exploratory; interpret "context-dependent" calls cautiously. - **How to assess confidence**: use this table primarily for hypothesis generation or to find lncRNAs associated with genes that already have strong methylation–miRNA context signals.- **multi_way_interactions.csv** (if multi-way module is enabled): - Summarizes multi-regulator regression models for each gene, comparing a simple model to a full model with many regulators. - Contains `improvement_from_regulators` and an F-test `interaction_p_value` with a boolean `has_significant_interactions`. - **How to assess confidence**: current evidence shows that large improvements can also arise in randomized data; treat these results as exploratory and consider additional validation (e.g., overlap with simpler context metrics or external datasets).## Analysis Parameters- **Parallel workers:** 192- **Data directory:** /mmfs1/gscratch/scrubbed/sr320/github/ConTra/data/full-species-biomin/cleaned_ptua- **Output directory:** /mmfs1/gscratch/scrubbed/sr320/github/ConTra/output/context_dependent_analysis_20251201_160901- **Analysis timestamp:** 20251201_160901## ConclusionThis analysis successfully identified context-dependent regulatory interactions using optimized parallel processing.The results provide insights into how different regulatory layers interact in a context-specific manner.*Report generated by OptimizedContextDependentRegulationAnalysis on 2025-12-02 12:52:16*# Peve Context-Dependent Regulation Analysis Report**Generated:** 2025-11-30 19:41:04**Analysis ID:** 20251130_091333## Executive SummaryThis report presents the results of context-dependent regulatory interaction analysis between:- Gene expression- miRNA expression- lncRNA expression- DNA methylation## Analysis Overview- **Parallel Processing:** 192 CPU cores- **Available RAM:** 2930.5 GB- **Datasets Loaded:** 4 data types - Gene: 36 samples × 553 features - Lncrna: 36 samples × 8319 features - Mirna: 36 samples × 48 features - Methylation: 36 samples × 104459 features## Results Summary### Methylation-miRNA Context Analysis- **Total interactions analyzed:** 27150- **Context-dependent interactions (F-test):** 3935- **Mean improvement from interaction:** 0.022- **Mean context strength:** 0.371#### Top 10 Methylation-miRNA Interactions (by Context Strength)*This table shows genes whose regulation by DNA methylation is context-dependent on miRNA expression levels, ranked by context strength.*| Rank | Gene | Methylation | miRNA | Improvement | Context Strength | Empirical FDR Sig. ||------|------|-------------|-------|-------------|------------------|--------------------|| 1 | ('OG_11813', 'Peve_00021937') | methylation_CpG_Porites_evermani_scaffold_3933_26651 | mirna_Cluster_1004 | 0.028 | 1.523 | False || 2 | ('OG_11813', 'Peve_00021937') | methylation_CpG_Porites_evermani_scaffold_2342_55729 | mirna_Cluster_1004 | 0.043 | 1.513 | False || 3 | ('OG_11813', 'Peve_00021937') | methylation_CpG_Porites_evermani_scaffold_3933_26651 | mirna_Cluster_6236 | 0.052 | 1.455 | False || 4 | ('OG_08793', 'Peve_00007107') | methylation_CpG_Porites_evermani_scaffold_865_27253 | mirna_Cluster_3165 | 0.032 | 1.451 | False || 5 | ('OG_11813', 'Peve_00021937') | methylation_CpG_Porites_evermani_scaffold_3933_26651 | mirna_Cluster_4247 | 0.038 | 1.437 | False || 6 | ('OG_11360', 'Peve_00001484') | methylation_CpG_Porites_evermani_scaffold_325_68647 | mirna_Cluster_6234 | 0.130 | 1.388 | False || 7 | ('OG_06599', 'Peve_00038759') | methylation_CpG_Porites_evermani_scaffold_259_87918 | mirna_Cluster_9197 | 0.000 | 1.369 | False || 8 | ('OG_11360', 'Peve_00001484') | methylation_CpG_Porites_evermani_scaffold_325_68647 | mirna_Cluster_3165 | 0.164 | 1.362 | False || 9 | ('OG_11813', 'Peve_00021937') | methylation_CpG_Porites_evermani_scaffold_1152_147508 | mirna_Cluster_1004 | 0.054 | 1.360 | False || 10 | ('OG_07153', 'Peve_00000682') | methylation_CpG_Porites_evermani_scaffold_910_155276 | mirna_Cluster_5058 | 0.078 | 1.358 | False |### lncRNA-miRNA Context Analysis (Exploratory)- **Total interactions analyzed:** 27150- **Context-dependent interactions (F-test):** 5550- **Mean improvement from interaction:** 0.029- **Mean context strength:** 0.282#### Top 10 lncRNA-miRNA Interactions (Exploratory)*This table identifies genes whose lncRNA-mediated regulation varies depending on miRNA expression. Note that current interaction metrics behave similarly or more strongly under randomized data and should be interpreted cautiously.*| Rank | Gene | lncRNA | miRNA | Improvement | Context Strength ||------|------|--------|-------|-------------|------------------|| 1 | ('OG_18262', 'Peve_00044240') | lncrna_lncRNA_10659 | mirna_Cluster_6099 | 0.469 | 1.007 || 2 | ('OG_18262', 'Peve_00044240') | lncrna_lncRNA_10659 | mirna_Cluster_6918 | 0.458 | 0.989 || 3 | ('OG_13684', 'Peve_00018981') | lncrna_lncRNA_12581 | mirna_Cluster_6234 | 0.444 | 0.898 || 4 | ('OG_12320', 'Peve_00024570') | lncrna_lncRNA_19767 | mirna_Cluster_670 | 0.440 | nan || 5 | ('OG_12320', 'Peve_00024570') | lncrna_lncRNA_19765 | mirna_Cluster_670 | 0.439 | nan || 6 | ('OG_13526', 'Peve_00019641') | lncrna_lncRNA_20807 | mirna_Cluster_670 | 0.437 | nan || 7 | ('OG_13526', 'Peve_00019641') | lncrna_lncRNA_23227 | mirna_Cluster_670 | 0.437 | nan || 8 | ('OG_13684', 'Peve_00018981') | lncrna_lncRNA_23131 | mirna_Cluster_981 | 0.433 | 0.861 || 9 | ('OG_13684', 'Peve_00018981') | lncrna_lncRNA_12581 | mirna_Cluster_5257 | 0.433 | 0.975 || 10 | ('OG_13684', 'Peve_00018981') | lncrna_lncRNA_12581 | mirna_Cluster_6235 | 0.432 | 0.987 |### Multi-Way Interaction Analysis (Exploratory)- **Total genes analyzed:** 553- **Genes with significant interactions (F-test):** 0- **Mean improvement from interactions:** 0.574#### Top 10 Multi-Way Interactions (Exploratory)*This table shows genes with the largest multi-regulator improvements. Current evidence suggests similar improvements can arise under randomized data; treat these results as hypothesis-generating rather than definitive.*| Rank | Gene | Improvement | Significant Interactions ||------|------|-------------|------------------------|| 1 | ('OG_13066', 'Peve_00003034') | 0.921 | False || 2 | ('OG_13685', 'Peve_00034973') | 0.921 | False || 3 | ('OG_10907', 'Peve_00033654') | 0.921 | False || 4 | ('OG_05950', 'Peve_00044664') | 0.921 | False || 5 | ('OG_01981', 'Peve_00033650') | 0.921 | False || 6 | ('OG_03267', 'Peve_00013805') | 0.920 | False || 7 | ('OG_17015', 'Peve_00023327') | 0.919 | False || 8 | ('OG_02088', 'Peve_00026747') | 0.919 | False || 9 | ('OG_05713', 'Peve_00041095') | 0.919 | False || 10 | ('OG_12805', 'Peve_00005899') | 0.919 | False |## Data Files<https://gannet.fish.washington.edu/v1_web/owlshell/bu-github/ConTra/output/context_dependent_analysis_20251130_091333/tables/>The following data files were generated:These CSV files contain the detailed results of context-dependent regulatory analysis, including interaction statistics, p-values, context strengths, and regulatory relationships for each analyzed gene-regulator pair.- **high_methylation_gene_lncrna_correlations.csv** (69461.9 KB)- **high_methylation_gene_methylation_correlations.csv** (675831.9 KB)- **high_methylation_gene_mirna_correlations.csv** (262.4 KB)- **high_mirna_gene_lncrna_correlations.csv** (65272.2 KB)- **high_mirna_gene_methylation_correlations.csv** (604621.8 KB)- **high_mirna_gene_mirna_correlations.csv** (144.7 KB)- **lncrna_mirna_context.csv** (6740.5 KB)- **low_mirna_gene_lncrna_correlations.csv** (111381.4 KB)- **low_mirna_gene_methylation_correlations.csv** (641498.3 KB)- **low_mirna_gene_mirna_correlations.csv** (171.5 KB)- **methylation_mirna_context.csv** (7651.1 KB)- **multi_way_interactions.csv** (1213.2 KB)### Table Definitions and Statistical ConfidenceBelow is a brief description of the main table types and how to assess statistical confidence for each:- **methylation_mirna_context.csv**: - One row per gene–CpG–miRNA triplet. - Contains regression-based metrics (`r2_*`, `improvement_from_regulator2`, `improvement_from_interaction`), conditional correlations (`corr_high_regulator2`, `corr_low_regulator2`), and `context_strength`. - When empirical FDR is enabled, includes `empirical_fdr_threshold`, `empirical_fdr_estimated`, and `empirical_fdr_significant`. - **How to assess confidence**: prioritize interactions with `empirical_fdr_significant == True` (if available); otherwise, use `context_strength` as the primary effect-size metric, ideally cross-referenced against random-data null runs.- **high_methylation_gene_methylation_correlations.csv** / **high_methylation_gene_mirna_correlations.csv** / **high_methylation_gene_lncrna_correlations.csv**: - Correlation networks in the **high-methylation context**, defined using a sentinel CpG site. - Each row is a gene–regulator pair with a Pearson correlation (`correlation`) and raw `p_value` computed only within high-methylation samples. - **How to assess confidence**: these p-values are not corrected for multiple testing; treat them as exploratory. For higher confidence, focus on strong effect sizes (|correlation| close to 1) and/or overlap with FDR-supported context interactions from `methylation_mirna_context.csv`.- **low_mirna_gene_methylation_correlations.csv** / **low_mirna_gene_mirna_correlations.csv** / **low_mirna_gene_lncrna_correlations.csv**: - Correlation networks in the **low-miRNA context**, using a sentinel miRNA as the context-defining variable. - Each row is a gene–regulator pair with a Pearson correlation (`correlation`) and raw `p_value` computed only within low-miRNA samples. - **How to assess confidence**: as above, p-values are unadjusted across many tests; use them as a guide for ranking, not strict significance. Strong |correlation| and consistency with patterns seen in `methylation_mirna_context.csv` or across contexts provide more persuasive evidence.- **lncrna_mirna_context.csv** (if lncRNA context module is enabled): - Regression-based lncRNA–miRNA–gene context metrics analogous to `methylation_mirna_context.csv`. - Empirical comparisons with randomized data suggest that current interaction metrics here are more exploratory; interpret "context-dependent" calls cautiously. - **How to assess confidence**: use this table primarily for hypothesis generation or to find lncRNAs associated with genes that already have strong methylation–miRNA context signals.- **multi_way_interactions.csv** (if multi-way module is enabled): - Summarizes multi-regulator regression models for each gene, comparing a simple model to a full model with many regulators. - Contains `improvement_from_regulators` and an F-test `interaction_p_value` with a boolean `has_significant_interactions`. - **How to assess confidence**: current evidence shows that large improvements can also arise in randomized data; treat these results as exploratory and consider additional validation (e.g., overlap with simpler context metrics or external datasets).## Analysis Parameters- **Parallel workers:** 192- **Data directory:** /mmfs1/gscratch/scrubbed/sr320/github/ConTra/data/full-species-biomin/cleaned_peve- **Output directory:** /mmfs1/gscratch/scrubbed/sr320/github/ConTra/output/context_dependent_analysis_20251130_091333- **Analysis timestamp:** 20251130_091333## ConclusionThis analysis successfully identified context-dependent regulatory interactions using optimized parallel processing.The results provide insights into how different regulatory layers interact in a context-specific manner.*Report generated by OptimizedContextDependentRegulationAnalysis on 2025-11-30 19:41:04*# Apul Context-Dependent Regulation Analysis Report**Generated:** 2025-12-01 11:13:07**Analysis ID:** 20251201_061836## Executive SummaryThis report presents the results of context-dependent regulatory interaction analysis between:- Gene expression- miRNA expression- lncRNA expression- DNA methylation## Analysis Overview- **Parallel Processing:** 192 CPU cores- **Available RAM:** 2940.2 GB- **Datasets Loaded:** 4 data types - Gene: 39 samples × 517 features - Lncrna: 39 samples × 15559 features - Mirna: 39 samples × 51 features - Methylation: 39 samples × 66428 features## Results Summary### Methylation-miRNA Context Analysis- **Total interactions analyzed:** 51150- **Context-dependent interactions (F-test):** 8765- **Mean improvement from interaction:** 0.027- **Mean context strength:** 0.346#### Top 10 Methylation-miRNA Interactions (by Context Strength)*This table shows genes whose regulation by DNA methylation is context-dependent on miRNA expression levels, ranked by context strength.*| Rank | Gene | Methylation | miRNA | Improvement | Context Strength | Empirical FDR Sig. ||------|------|-------------|-------|-------------|------------------|--------------------|| 1 | ('OG_05303', 'FUN_023974') | methylation_CpG_ptg000001l_9710125 | mirna_Cluster_3109 | 0.060 | 1.881 | False || 2 | ('OG_07265', 'FUN_033056') | methylation_CpG_ptg000021l_10139814 | mirna_Cluster_3109 | 0.018 | 1.875 | False || 3 | ('OG_14547', 'FUN_019026') | methylation_CpG_ptg000023l_11736283 | mirna_Cluster_3109 | 0.023 | 1.875 | False || 4 | ('OG_07265', 'FUN_033056') | methylation_CpG_ptg000018l_7377398 | mirna_Cluster_3109 | 0.067 | 1.854 | False || 5 | ('OG_10509', 'FUN_002531') | methylation_CpG_ptg000031l_5310815 | mirna_Cluster_3109 | 0.004 | 1.837 | False || 6 | ('OG_06610', 'FUN_029760') | methylation_CpG_ptg000031l_5310762 | mirna_Cluster_3109 | 0.004 | 1.833 | False || 7 | ('OG_10811', 'FUN_006888') | methylation_CpG_ptg000023l_8252944 | mirna_Cluster_3109 | 0.006 | 1.756 | False || 8 | ('OG_14711', 'FUN_024326') | methylation_CpG_ptg000012l_17885415 | mirna_Cluster_3109 | 0.045 | 1.672 | False || 9 | ('OG_10811', 'FUN_006888') | methylation_CpG_ptg000027l_7575122 | mirna_Cluster_3109 | 0.015 | 1.660 | False || 10 | ('OG_06610', 'FUN_029760') | methylation_CpG_ptg000021l_6263529 | mirna_Cluster_3109 | 0.012 | 1.657 | False |### lncRNA-miRNA Context Analysis (Exploratory)- **Total interactions analyzed:** 51150- **Context-dependent interactions (F-test):** 8365- **Mean improvement from interaction:** 0.021- **Mean context strength:** 0.270#### Top 10 lncRNA-miRNA Interactions (Exploratory)*This table identifies genes whose lncRNA-mediated regulation varies depending on miRNA expression. Note that current interaction metrics behave similarly or more strongly under randomized data and should be interpreted cautiously.*| Rank | Gene | lncRNA | miRNA | Improvement | Context Strength ||------|------|--------|-------|-------------|------------------|| 1 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_48837 | mirna_Cluster_14165 | 0.420 | 1.162 || 2 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_48837 | mirna_Cluster_14146 | 0.407 | 1.082 || 3 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_48837 | mirna_Cluster_3226 | 0.406 | 0.969 || 4 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_48837 | mirna_Cluster_10452 | 0.400 | 0.910 || 5 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_23415 | mirna_Cluster_9512 | 0.394 | 1.033 || 6 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_23414 | mirna_Cluster_9512 | 0.392 | 1.021 || 7 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_23416 | mirna_Cluster_9512 | 0.391 | 1.028 || 8 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_54028 | mirna_Cluster_4752 | 0.387 | 0.966 || 9 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_48837 | mirna_Cluster_4752 | 0.379 | 1.288 || 10 | ('OG_10579', 'FUN_003891') | lncrna_lncRNA_44695 | mirna_Cluster_4752 | 0.377 | 0.834 |### Multi-Way Interaction Analysis (Exploratory)- **Total genes analyzed:** 517- **Genes with significant interactions (F-test):** 0- **Mean improvement from interactions:** 0.750#### Top 10 Multi-Way Interactions (Exploratory)*This table shows genes with the largest multi-regulator improvements. Current evidence suggests similar improvements can arise under randomized data; treat these results as hypothesis-generating rather than definitive.*| Rank | Gene | Improvement | Significant Interactions ||------|------|-------------|------------------------|| 1 | ('OG_10450', 'FUN_001498') | 0.928 | False || 2 | ('OG_12424', 'FUN_030063') | 0.928 | False || 3 | ('OG_10523', 'FUN_002672') | 0.928 | False || 4 | ('OG_12286', 'FUN_028190') | 0.926 | False || 5 | ('OG_09897', 'FUN_042992') | 0.925 | False || 6 | ('OG_14500', 'FUN_017790') | 0.925 | False || 7 | ('OG_04209', 'FUN_016791') | 0.923 | False || 8 | ('OG_10759', 'FUN_006140') | 0.922 | False || 9 | ('OG_08920', 'FUN_038376') | 0.921 | False || 10 | ('OG_02176', 'FUN_008525') | 0.920 | False |## Data Files<https://gannet.fish.washington.edu/v1_web/owlshell/bu-github/ConTra/output/context_dependent_analysis_20251201_061836/tables/>The following data files were generated:These CSV files contain the detailed results of context-dependent regulatory analysis, including interaction statistics, p-values, context strengths, and regulatory relationships for each analyzed gene-regulator pair.- **high_methylation_gene_lncrna_correlations.csv** (130703.2 KB)- **high_methylation_gene_methylation_correlations.csv** (284276.8 KB)- **high_methylation_gene_mirna_correlations.csv** (177.7 KB)- **lncrna_mirna_context.csv** (13579.9 KB)- **low_mirna_gene_lncrna_correlations.csv** (96572.2 KB)- **low_mirna_gene_methylation_correlations.csv** (343631.8 KB)- **low_mirna_gene_mirna_correlations.csv** (266.4 KB)- **methylation_mirna_context.csv** (14423.3 KB)- **multi_way_interactions.csv** (968.1 KB)### Table Definitions and Statistical ConfidenceBelow is a brief description of the main table types and how to assess statistical confidence for each:- **methylation_mirna_context.csv**: - One row per gene–CpG–miRNA triplet. - Contains regression-based metrics (`r2_*`, `improvement_from_regulator2`, `improvement_from_interaction`), conditional correlations (`corr_high_regulator2`, `corr_low_regulator2`), and `context_strength`. - When empirical FDR is enabled, includes `empirical_fdr_threshold`, `empirical_fdr_estimated`, and `empirical_fdr_significant`. - **How to assess confidence**: prioritize interactions with `empirical_fdr_significant == True` (if available); otherwise, use `context_strength` as the primary effect-size metric, ideally cross-referenced against random-data null runs.- **high_methylation_gene_methylation_correlations.csv** / **high_methylation_gene_mirna_correlations.csv** / **high_methylation_gene_lncrna_correlations.csv**: - Correlation networks in the **high-methylation context**, defined using a sentinel CpG site. - Each row is a gene–regulator pair with a Pearson correlation (`correlation`) and raw `p_value` computed only within high-methylation samples. - **How to assess confidence**: these p-values are not corrected for multiple testing; treat them as exploratory. For higher confidence, focus on strong effect sizes (|correlation| close to 1) and/or overlap with FDR-supported context interactions from `methylation_mirna_context.csv`.- **low_mirna_gene_methylation_correlations.csv** / **low_mirna_gene_mirna_correlations.csv** / **low_mirna_gene_lncrna_correlations.csv**: - Correlation networks in the **low-miRNA context**, using a sentinel miRNA as the context-defining variable. - Each row is a gene–regulator pair with a Pearson correlation (`correlation`) and raw `p_value` computed only within low-miRNA samples. - **How to assess confidence**: as above, p-values are unadjusted across many tests; use them as a guide for ranking, not strict significance. Strong |correlation| and consistency with patterns seen in `methylation_mirna_context.csv` or across contexts provide more persuasive evidence.- **lncrna_mirna_context.csv** (if lncRNA context module is enabled): - Regression-based lncRNA–miRNA–gene context metrics analogous to `methylation_mirna_context.csv`. - Empirical comparisons with randomized data suggest that current interaction metrics here are more exploratory; interpret "context-dependent" calls cautiously. - **How to assess confidence**: use this table primarily for hypothesis generation or to find lncRNAs associated with genes that already have strong methylation–miRNA context signals.- **multi_way_interactions.csv** (if multi-way module is enabled): - Summarizes multi-regulator regression models for each gene, comparing a simple model to a full model with many regulators. - Contains `improvement_from_regulators` and an F-test `interaction_p_value` with a boolean `has_significant_interactions`. - **How to assess confidence**: current evidence shows that large improvements can also arise in randomized data; treat these results as exploratory and consider additional validation (e.g., overlap with simpler context metrics or external datasets).## Analysis Parameters- **Parallel workers:** 192- **Data directory:** /mmfs1/gscratch/scrubbed/sr320/github/ConTra/data/full-species-biomin/cleaned_apul- **Output directory:** /mmfs1/gscratch/scrubbed/sr320/github/ConTra/output/context_dependent_analysis_20251201_061836- **Analysis timestamp:** 20251201_061836## ConclusionThis analysis successfully identified context-dependent regulatory interactions using optimized parallel processing.The results provide insights into how different regulatory layers interact in a context-specific manner.*Report generated by OptimizedContextDependentRegulationAnalysis on 2025-12-01 11:13:07*