Here are few tidbits to bring you cheer this Christmas Season!
In no particular order:
1) [Paper] Genomic insights into the origin of ecotypes
Highlights
The ecotype concept describes populations of species that are phenotypically and genetically differentiated by adaptation to contrasting habitats.
Ecotypes offer a powerful framework for understanding how, and why, biodiversity arises within species and why similar environments repeatedly generate similar forms.
A century after its introduction, the ecotype concept continues to shape thinking about adaptation, ecological diversification, and the early stages of speciation.
The concept has expanded far beyond its botanical origins, influencing research across the tree of life.
Genomic tools have provided new insight into the forces and factors that drive the formation of ecotypes.
Integrating genomic, ecological, and experimental approaches will be essential for tackling emerging challenges in the decade ahead.
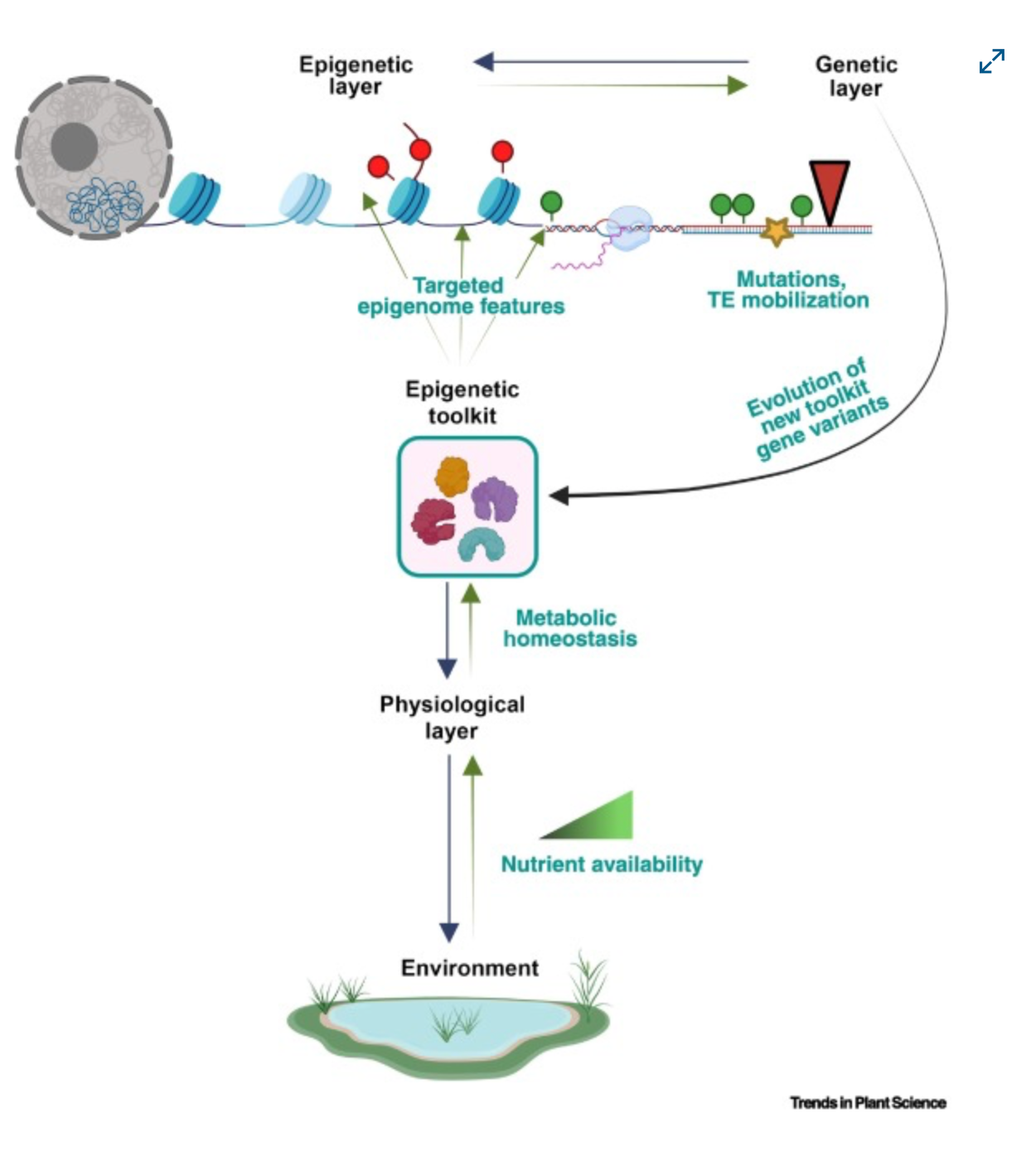
2) [Paper] Defining the epigenetic toolkit as an evolvable trait
Highlights
Previous studies have revealed the genetic basis of many complex epigenetic and phenotypic traits through classic screening or genome-wide association studies (GWAS). We explore the question of how the homeostatic state of a plant is balanced under varied levels of nutrient supply when environmental or internal (epi)genetic factors significantly perturb this balance.
We explore how epigenetic regulation of the genome of a plant interacts with physiological processes (such as homeostasis) at the biological pathway level when exposed to different nutrient availability scenarios and under changing environmental conditions.
We propose a conceptual model involving an ‘epigenetic toolkit’ where plants sense environmental pressures to develop a complex crosstalk between physiology, epigenetic/epigenomic state, and their genetic code, and ultimately fine-tune adaptation at the organismal level.
3) [Software] IsoformSwitchAnalyzeR v2: Analysis of Functional Isoform Changes in Long-read and Single-cell Sequencing Data
4) [Book] Modern Statistics for Modern Biology
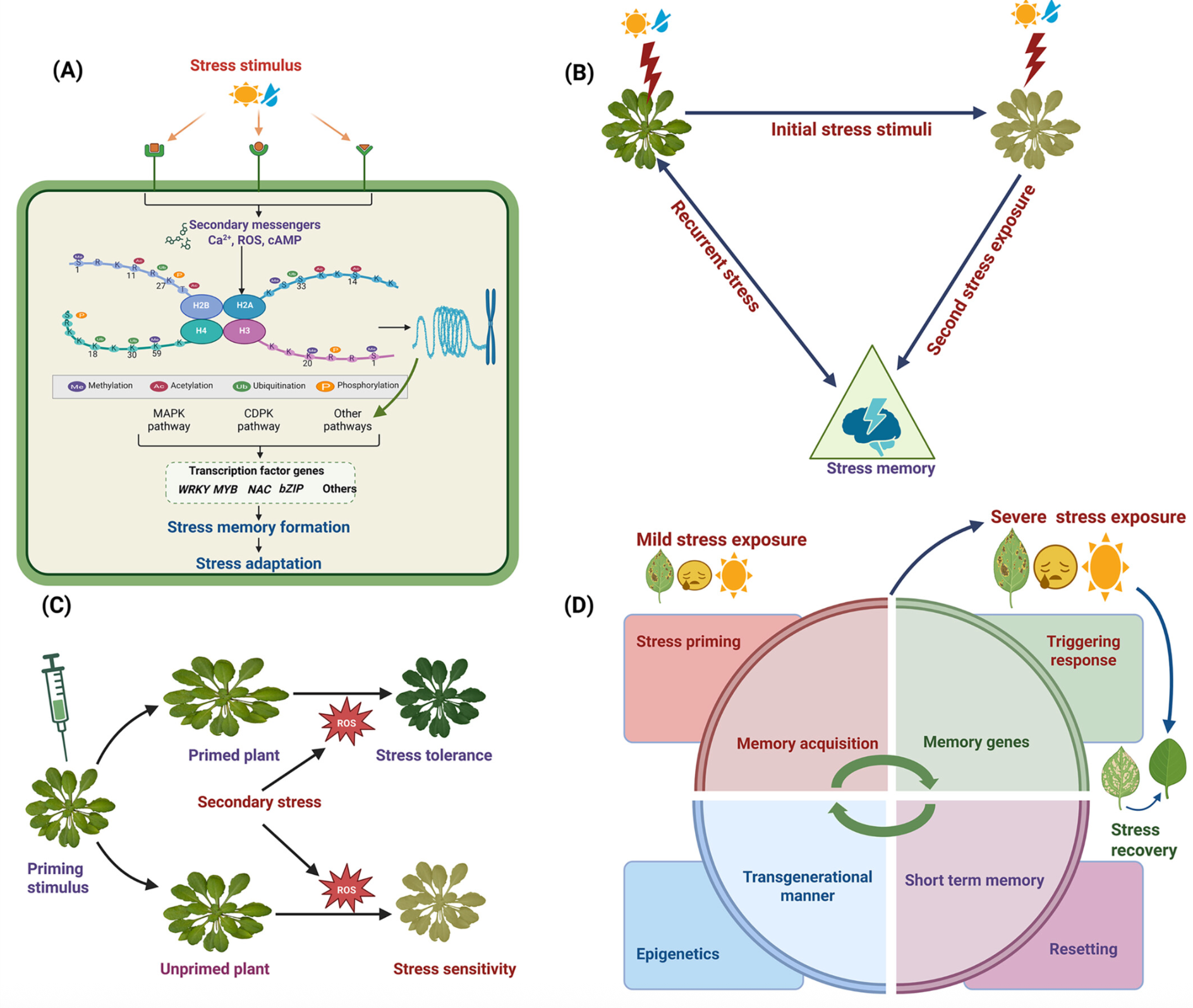
5) [Paper] The Plant Mind: Unraveling Abiotic Stress Priming, Memory, and Adaptation
ABSTRACT
Plants exhibit a remarkable capacity to adapt to recurrent abiotic stresses, prompting a re-evaluation of traditional views on plant responses to environmental challenges. This review explores the intricate mechanisms of stress priming, memory, and adaptation in plants. Specifically, it details the molecular and physiological processes underlying abiotic stress priming, which serve as a gateway to understanding plant memory. Stress priming fosters resilience against diverse stressors through interconnected pathways involving hormone signaling, transcriptional regulation, DNA methylation, histone modifications, and small RNAs. These epigenetic changes orchestrate stress-responsive gene expression and can, in some cases, be passed on to future generations. This review distinguishes between somatic memory, intergenerational effects, and transgenerational inheritance to avoid conceptual overlap. By connecting short-term priming to long-term adaptation and potential heritability, this article proposes a paradigm shift in how plant resilience is understood, with significant implications for crop improvement under climate stress.
6) [Software] GFF Analysis Tools - AI Agent
This AI agent provides an intuitive interface for bioinformatics researchers to analyze GFF files without writing complex code. Users can ask questions in natural language, and the agent will select and execute the appropriate analysis tools to provide comprehensive answers.
Features
🧬 Coordinate-based Queries
- Find features by genomic coordinates (regions and specific positions)
- Query features overlapping genomic regions with filtering by type and strand
- Identify features containing specific genomic positions
🔗 Relationship and Hierarchy Analysis
- Explore gene structure and organization (get all child features like exons, CDS, UTRs)
- Find parent features of any given feature using upward traversal
- Get all features of specific types with efficient iteration
📊 Statistical Analysis
- Calculate comprehensive feature statistics (counts, length distributions per feature type)
- Generate per-chromosome feature summaries and analysis
- Analyze length distributions with detailed statistics (min, max, mean, median, std dev, percentiles)
- Create histogram data for feature length distributions
🔍 Attribute-based Searches
- Search features by attribute key-value pairs (exact and partial matching)
- Find features containing specific attribute keys
- Support pattern matching and logical operations for attribute queries
📍 Positional Analysis
- Identify intergenic regions (gaps between genes) with filtering options
- Calculate feature density in genomic windows across chromosomes
- Analyze strand distribution of features with counts and percentages
- Support clustering analysis and positional insights
📤 Export and Reporting
- Export feature data to CSV format with comprehensive filtering
- Generate human-readable summary reports of GFF file contents
- Provide formatted output for downstream analysis
7) [Paper] Comprehensive whole transcriptome analysis reveals specific lncRNA, circRNA and miRNA-mRNA networks of pearl oyster (Pinctada fucata martensii) in response to hypoxia stress
Highlights
Construct a ceRNA network with 9 DElncRNAs, 10 DEcircRNAs, 3 DEmiRNAs, and 6 DEmRNAs.
CeRNA networks are critically involved in the molecular response to hypoxia.
Hypoxia drives ceRNA-based regulation of metabolism, antioxidation, and RNA splicing.
Abstract
Hypoxia is a prevalent environmental stressor in both natural and aquaculture environments. Pinctada fucata martensii, a commercially valuable pearl oyster species, is particularly susceptible to hypoxic stress in aquaculture. To explore its molecular response to short-term hypoxia, whole-transcriptome sequencing was performed on gill tissues under normoxic and hypoxic conditions. A total of 721 differentially expressed mRNAs (DEmRNAs), 259 long non-coding RNAs (DElncRNAs), 55 circular RNAs (DEcircRNAs), and 17 microRNAs (DEmiRNAs) were identified. Based on predicted interactions among these differentially expressed RNAs, a competing endogenous RNA (ceRNA) network was constructed, consisting of 9 DElncRNAs, 10 DEcircRNAs, 3 DEmiRNAs, and 6 DEmRNAs. Six DElncRNAs and three DEcircRNAs were predicted to interact with miR-152, and the axis-mediated activation of target genes–ALDH5A1 and SLC23A2, FAT4 and CCDC97, and LAC–was implicated in the modulation of metabolic pathways, enzyme activity, antioxidant defenses, thereby promoting antioxidant capacity and suppressing cell growth as part of an adaptive survival strategy. This ceRNA network reveals a key molecular hub that links hypoxia sensing to immunomodulatory mechanisms. Moreover, the co-repression of stress-response genes HSP20 and HSP70 in this network enhances hypoxic tolerance increased energy utilization efficiency and regulation of RNA. Functional analysis of the network revealed enrichment in pathways related to energy metabolism, antioxidant defense, and RNA splicing regulation. These findings provide novel insights into transcriptomic regulation and adaptive mechanisms in bivalves under hypoxic stress.
8 [Paper] Multi-Omics Reprogramming Drives a Counterintuitive Reversal of Disease Susceptibility During Ageing
Abstract
Ageing is a progressive and irreversible biological process characterized by the deterioration of physiological functions and increased vulnerability to mortality. Although extensively studied in vertebrates, ageing in long-lived invertebrates remains comparatively unexplored. While ageing typically leads to greater susceptibility to infectious diseases, a striking and unexpected reversal was identified in oysters: older oysters exhibit markedly increased tolerance to the Pacific Oyster Mortality Syndrome (POMS), a panzootic disease primarily driven by the OsHV-1 herpesvirus and responsible for severe losses in global aquaculture. To investigate this counterintuitive pattern, we challenged oysters aged 4, 16, and 28 months from four biparental families and conducted an integrative multi-omics analysis, including epigenomics, transcriptomics, and metabolomics on the two families showing the strongest age-related increase in survival. Our results reveal that ageing in Magallana gigas is characterized by coordinated epigenetic, transcriptional, and metabolic reprogramming that reduces host permissiveness to POMS. We show that the epigenetic remodeling of key immune regulators (e.g., Toll-like receptors, MyD88) aligns with transcriptional rewiring of NF-κB and ubiquitin pathways, producing a finely tuned innate immune state marked by enhanced antiviral activity but reduced antibacterial responsiveness. We also identify age-related repression of mTOR signaling, likely promoting autophagy and improving viral control. These regulatory changes are tightly linked to metabolic adjustments, including reduced TCA cycle flux, remodeled nitrogen metabolism, and altered glutathione dynamics, which collectively support a stress-tolerant, energy-conserving phenotype. Together, our findings reveal a fundamental evolutionary trade-off: juveniles prioritize growth at the cost of viral susceptibility, whereas adults invest in cellular maintenance and antiviral preparedness.
9 [Paper] Unraveling the microRNA regulatory network underlying the planktonic-to-benthic transition in the Pacific oyster
Abstract
The Pacific oyster, Crassostrea gigas, undergoes a critical developmental transition from planktonic larvae to sessile benthic juveniles, a process essential for both aquaculture productivity and the stability of marine ecosystems. While environmental and neuroendocrine cues regulating this transition have been extensively studied, the contribution of microRNAs (miRNAs) remains largely unexplored. In this study, we employed a high-resolution temporal sampling strategy to investigate oyster metamorphosis, identified 33 miRNAs associated with larval settlement and metamorphosis. Additionally, we uncovered 35 highly correlated miRNA-target gene pairs, revealing a complex post-transcriptional regulatory network. We further validated the expression pattern of the potential regulatory hub miRNA novel_mir_80 and confirmed its presence through whole-mount in situ hybridization using LNA-modified probes. Functional annotation of target genes highlighted key roles in adhesion, signal transduction, protein homeostasis, cytoskeleton remodeling, genome stability, and transcriptional and epigenetic regulation. These results underscore the pivotal role of miRNAs in orchestrating developmental reprogramming during the planktonic-benthic transition. Our findings not only expand current understanding of miRNA-mediated gene regulation in C. gigas, but also provide novel molecular targets with potential applications in oyster breeding and aquaculture management.