Summary
For our tutorial this week, I read the paper assigned to everyone, a Roberts lab special:
Roberts SB and Gavery MR (2012) Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates? Front. Physio. 2:116. doi: 10.3389/fphys.2011.00116
which reviewed previous research on DNA methylation and phenotypic plasticity in invertebrates, particularly non-model organisms such as the western honeybee and the Pacific oyster.
We were required to find our own, more recent review though, which led me to this paper:
Srut M (2021) Ecotoxicological epigenetics in invertebrates: Emerging tool for the evaluation of present and past pollution burden. Chemosphere. 282. doi: 10.1016/j.chemosphere.2021.131026
I will be giving summaries of both papers, comparing them, and describe an experiment that could integrate DNA methylation with my own interests in bivalve physiology.
Roberts and Gavery Review
This paper summarized current work looking to link DNA methylation with phenotypic plasticity in non-model invertebrates. In summarizing previous studies, Steven and Mackenzie found that where DNA is methylated and its degree of methylation varies by species. Moreover, in species with methylation (such as bees and oysters), functionally critical genes were originally hypothesized (and shown in computer analyses) to be more methylated than tissue-specific or inducible genes, and for that methylation to carry through the germline (the next generation). The rationale is that less methylation allows for greater transcriptional flexibility, which makes sense for genes that are constantly turning on and off, unlike genes that need to be expressed constantly (such as DNA and RNA metabolism). With less methylation, the gene can be more easily accessed by transcriptional machinery. This was confirmed by Steven and Mackenzie’s sequencing of methylated regions in the oyster genome, and a study done on western honeybees. See image below.
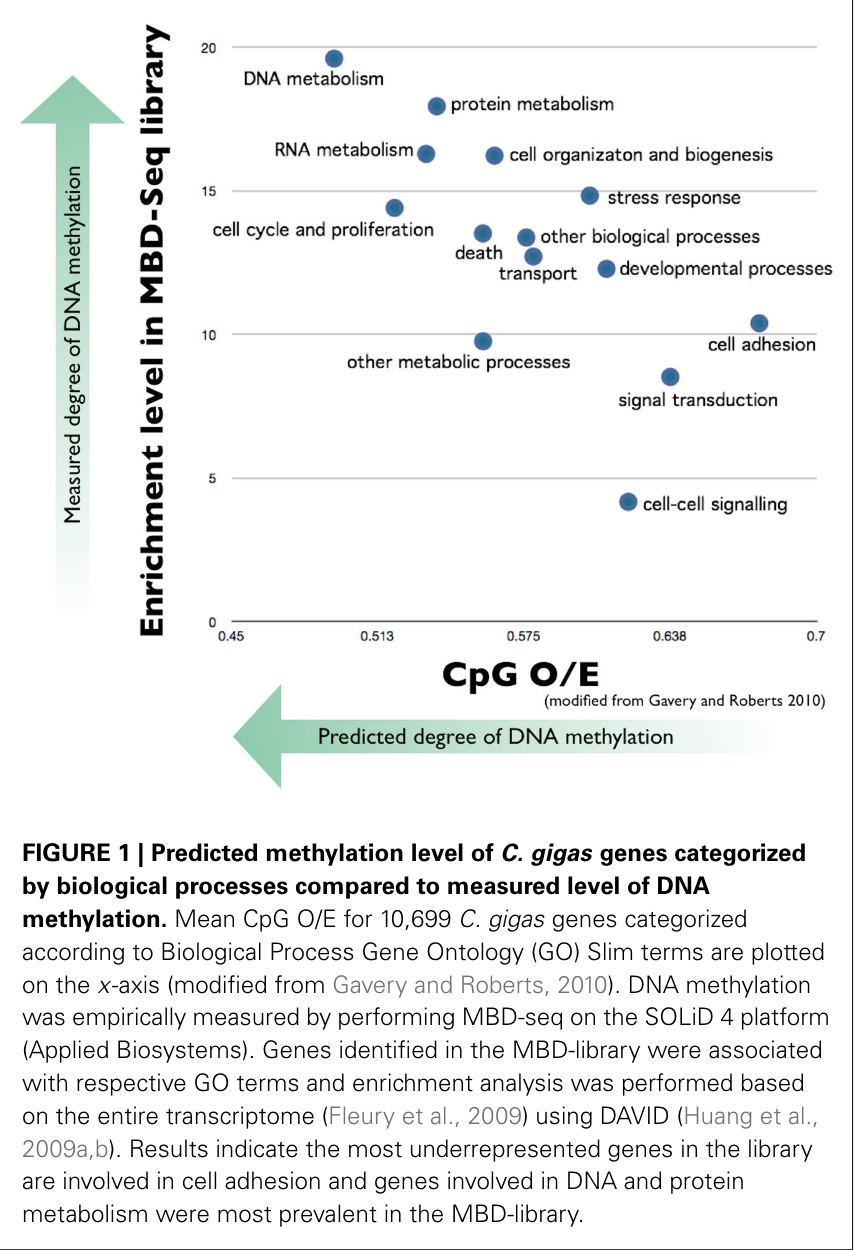
Methylation also varied depending on the presence of introns. Methylated genes also appeared to be more conserved across species, and unmethylated genes have more genetic diversity and theoretically more adaptive potential and phenotypic plasticity, via more or fewer alternative splicings of a certain gene, and other pathways.
Srut Review
This review focused on epigenetic modifications to invertebrate genomes following pollutant exposure, and the potential of using DNA methylation as a biomarker for pollution. The paper starts with a helpful review on what DNA methylation is and how it works before describing different studies of DNA methylation following pollutant exposure. The author then dives into how DNA methylation has changed in various invertebrates when exposed to metals, pesticides, harmful chemicals, radiation, BPA’s, etc. They include a three page table summarizing all the studies they could find. Next, they go into hypotheses of how pollutants alter methylation, with most of them involving hijacking or interruption of enzyme activity that carries out methylation. The next big chunk of the paper summarizes studies that have looked at DNA methylation after pollution over multiple generations after the original parent was exposed to a pollutant. Across multiple species and pollutants, they see mixes of hypo- and hypermethylation depending on the pollutant, the species, and the generation. The final part of the paper looks at challenges within the field of ecotoxicological epigenetics. Accessing robust bisulfite sequencing technology can often be prohibitively expensive, time-consuming, and heavily reliant on good expertise. Moreover, epigenetic data is often not linked to phenotypic expression within studies, and this is in part because of limited, well-characterized methylomes. The author also writes that in order for epigenetic data to be even relevant to ecotoxicology, the field must expand what organisms it uses so that it includes all ecologically relevant species, at different life stages, and all different tissues within an organism. The author concludes this section by saying that beyond DNA methylation, other gene regulatory (epigenetic) mechanisms need to be explored, such as non-coding RNA’s and histones.
Comparisons
| Similarities | Differences |
|---|---|
| -Agree that less DNA methylation is linked to increased plasticity -Both discussed DNA methylation in non-model organisms | -Srut is much newer (2021), talks about more organisms and techniques -Srut talked more specifically about pollutant research, Roberts more about phenotypic plasticity -Srut’s paper talked more about genome wide methylation, Roberts more about specific genes |
Applications to my research
I would love to do some targeted DNA methylation analysis in mussels that are exposed to high flow or low flow environments and see if that changes methylation in foot protein genes, given that mussels often lay more threads when exposed to higher flows. Although, that might not be super applicable, as we would likely need to see a change in the composition of the thread in order to make methylation analysis worthwhile. However, maybe there’s hypomethylation near the regulatory apparatus that controls byssal thread deposition. Could be interesting.