Paper I Chose
Long non-coding RNAs are associated with spatiotemporal gene expression profiles in the marine gastropod Tegula atra
Can find here
Background
Sampled gill tissue from snails located at 4 different sites along the central northern coast of Chile.
Gill tissue was sampled in summer and winter
Sequencing and lncRNA Identification
RNA-seq
lncRNA ID process filters for >200 bp, coding potential (CPAT), and BLAST against Genbank protein sequence database for mussels
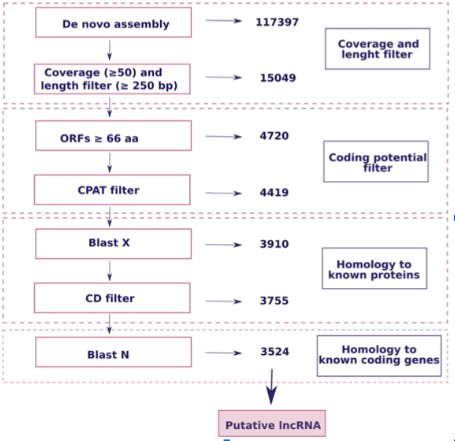
Analysis
Expression analysis for both lncRNAs and coding RNAs
PCA, differential expression, and expression correlation between lnc and coding RNAs
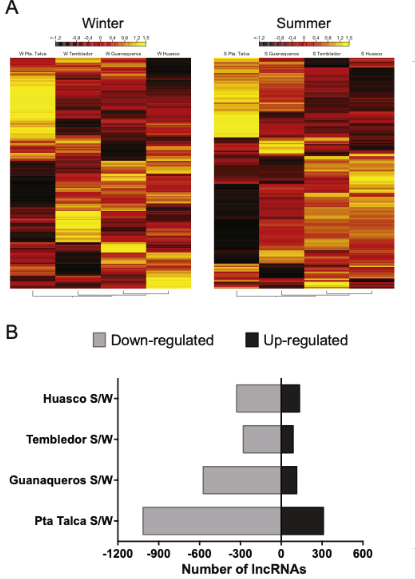
Differential expression of lncRNAs by season
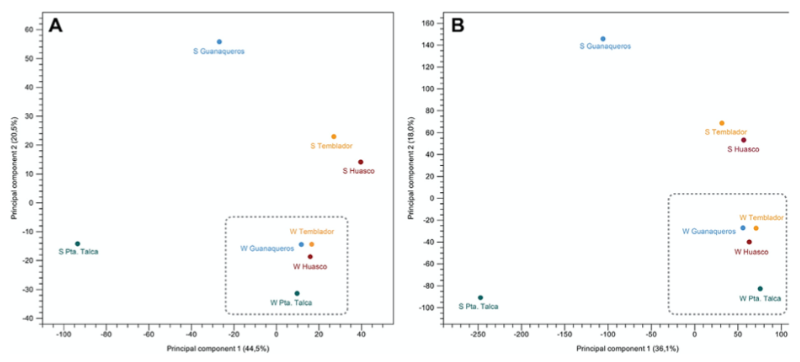
PCA
More variability in expression over summer, less in winter
Punta Talca individuals most distinct from others
Co-expression
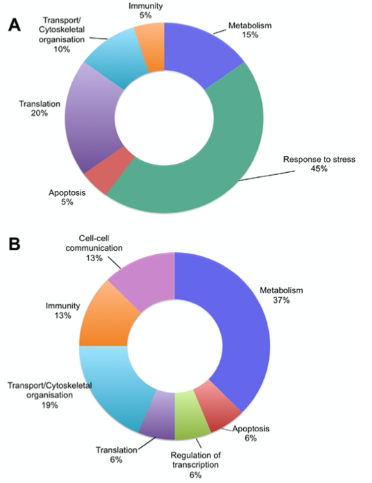
Correlation of lncRNA expression with known coding RNA expression
lncRNA upregulation (A) most associated with stress response
Downregulation (B) most associated with metabolism
My Main Takeaways
lncRNA expression showed seasonal variation as well as spatial variation–most notably in the case of a single site (thought to be due to more temperature variability at this site).
lncRNA expression appears to be very involved in stress response, translation, and metabolism–which play important roles during cases of environmental perturbation.
The author suggest that lncRNA genes may be more environmentally responsive than coding genes, implicating them more in rapid response to stress.