Geoduck RRBS- Locating 41
Sean has identified 41 loci that are different in the 3 treatments at Day 10!
I am going to try to see if I can find out where they are in the genome…
file: http://owl.fish.washington.edu/halfshell/working-directory/17-01-30/DiffMethRegions-41.bed
contents
scaffold103946 20422 20422 -0.1859
scaffold106982 14135 14135 1.451
scaffold107590 8577 8577 0.5295
scaffold112102 14447 14447 2.266
scaffold116821 17 17 0.6799
scaffold11815 14456 14456 3.786
scaffold11917 28699 28699 0.2738
scaffold1242 12404 12404 0.5895
scaffold126195 1416 1416 -1.394
scaffold128214 5365 5365 0.1436
scaffold129719 11544 11544 0.305
scaffold129719 11591 11591 0.3073
scaffold130369 15322 15322 0.2887
scaffold13704 29380 29380 -2.035
scaffold147711 174 174 0.2606
scaffold16116 7076 7076 -0.1679
scaffold176442 9109 9109 -2.641
scaffold180853 5067 5067 1.867
scaffold193328 3840 3840 2.474
scaffold20564 10594 10594 0.5333
scaffold21816 40741 40741 0.6428
scaffold21816 40765 40765 0.6775
scaffold26891 2783 2783 -0.9512
scaffold27468 17795 17795 0.4412
scaffold28099 1642 1642 0.4073
scaffold3069 4606 4606 -0.5935
scaffold30780 3832 3832 0.3615
scaffold318488 1576 1576 1.808
scaffold318488 1652 1652 6.167
scaffold3339 18557 18557 0.5385
scaffold3339 18559 18559 0.4022
scaffold420603 1339 1339 1.677
scaffold42112 6026 6026 -3.169
scaffold45108 21939 21939 5.189
scaffold5159 16185 16185 -0.4887
scaffold5906 3325 3325 -0.4556
scaffold59568 151 151 0.5148
scaffold62783 8660 8660 -0.73
scaffold69655 10128 10128 0.2736
scaffold8586 14161 14161 1.713
scaffold92878 9598 9598 -0.3131
I started an IGV session and realized hard drive failure required me to rerun some stuff..
First the DMR file needs to be adjusted so it the data falls over C - this should mean decreasing start position by one.
I have some IGV tracks from December (via early Bismark)
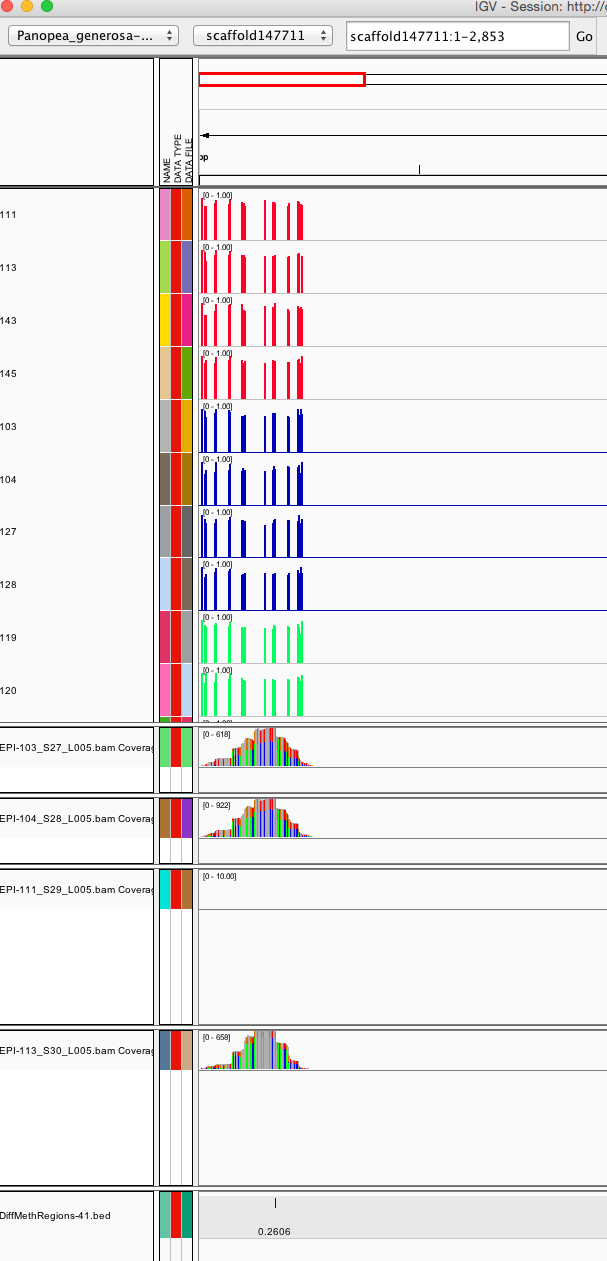
session file: http://owl.fish.washington.edu/halfshell/working-directory/17-01-30/igv_session-EPI-013017.xml
I have mapped some bam files for RRBS to see coverage.
Rerunning …
Crude mapping of RNA-seq to genome
https://github.com/sr320/nb-2017/blob/master/P_generosa/01-Mapping-RNA-seq.ipynb
Creating GFF of transcriptome blast
https://github.com/sr320/nb-2017/blob/master/P_generosa/02-Transcriptome-blastn.ipynb
