All gigas at once
What if we start with current data and worked our way back to see if an integration of data was fruitful. Step 1 - bismark it all together..
We have what we call Ronit’s data.
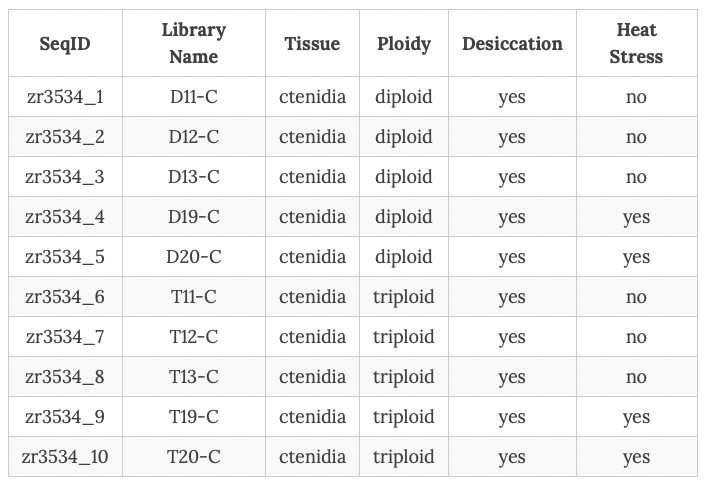
placing those samples in directory
/gscratch/srlab/sr320/data/cg-big/
specifically
cp /gscratch/srlab/mngeorge/data/cgigas_ronit/zr3534* .
We also have Roberto’s samples…
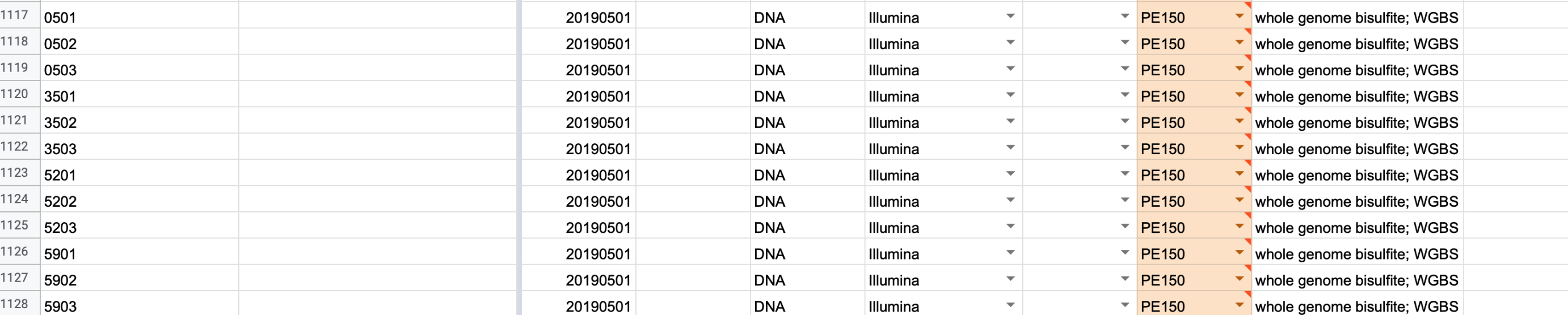
Moving trimmed files to mox.
wget -r --no-directories --no-parent -P . --no-check-certificate -A fastp-trim.20200819.fq.gz https://gannet.fish.washington.edu/Atumefaciens/20200818_cgig_wgbs_roberto_fastp_trimming/
Haws samples
3644 prefix
wget -r \
--no-directories --no-parent \
-P . \
--no-check-certificate \
-A "20201206.fq.gz" https://gannet.fish.washington.edu/Atumefaciens/20201206_cgig_fastp-10bp-5-3-prime_ploidy-pH-wgbs/
and
wget -r \
--no-directories --no-parent \
-P . \
--no-check-certificate \
-A "*val_*.fq.gz" https://gannet.fish.washington.edu/spartina/project-oyster-oa/Haws/trimmed-data-2/poly-G/
Manchester gonad
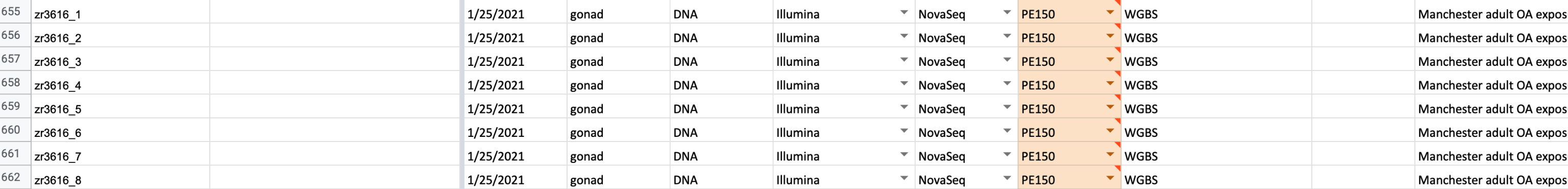
3616
wget -r \
--no-directories --no-parent \
-P . \
--no-check-certificate \
-A "*val_*.fq.gz" https://gannet.fish.washington.edu/spartina/project-gigas-oa-meth/output/trimgalore/03-poly-G/
Possibly some old samples?? maybe later
Claire and Yanouk..
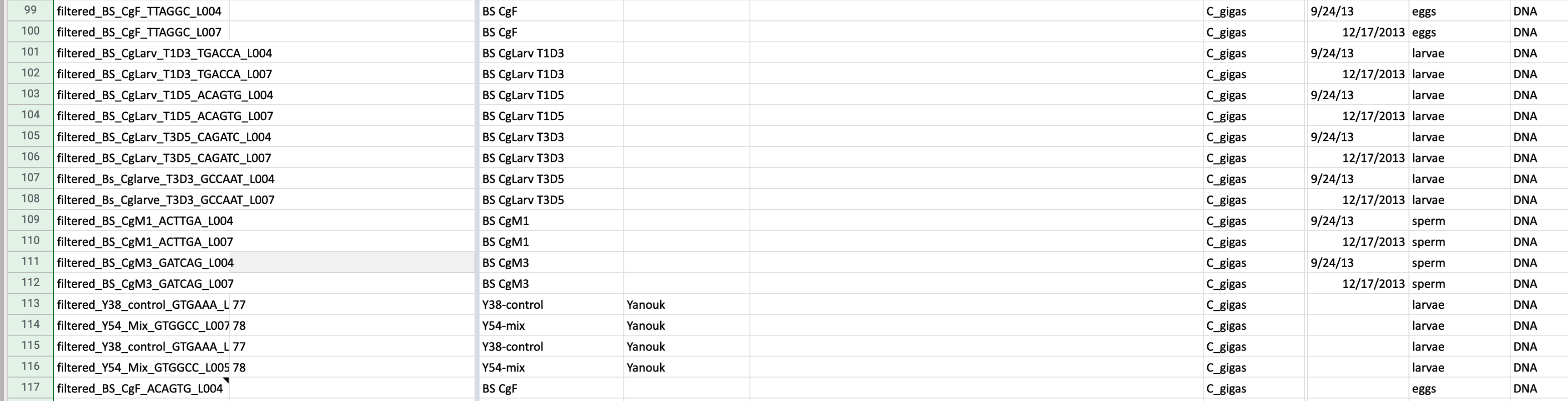
[sr320@mox1 cg-big]$ ls
0501_R1.fastp-trim.20200819.fq.gz zr3534_3_R2.fastp-trim.20201202.fq.gz zr3644_19_R1.fastp-trim.20201206.fq.gz
0501_R2.fastp-trim.20200819.fq.gz zr3534_4_R1.fastp-trim.20201202.fq.gz zr3644_19_R2.fastp-trim.20201206.fq.gz
0502_R1.fastp-trim.20200819.fq.gz zr3534_4_R2.fastp-trim.20201202.fq.gz zr3644_1_R1.fastp-trim.20201206.fq.gz
A little bit of renaming for fastp
for file in *.fq.gz ; do
mv $file $(echo $file | rev | cut -f2- -d- | rev | sed s/.fastp//g).fq.gz
done
after pulling down the trimgalore.. (Haws and Manchester) Want to save the duplicate Haws so
#zr3644_7_R2_val_2_val_2_val_2.fq.gz
for file in *R*_val* ; do
mv $file $(echo $file | rev | cut -f7- -d_ | rev | sed s/zr/zrtg/g).fq.gz
done
072121
Timed out at 20 days, missing about 5 samples. Will know those out in Bismark then combined all files to complete script.
Written on June 4, 2021
