Hemato qPCR in R
02-Crab-qpcr
library("dplyr")
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
qpcr <- read.csv("../data/hematqpcr_crabRNA.csv")
head(qpcr)
## Uniq_ID FRP tube_number trtmnt_tank sample_day infection_status maturity
## 1 6101_206_12 6101 206 cold 2 0 I
## 2 6101_415_26 6101 415 cold 17 0 I
## 3 6101_84_9 6101 84 <NA> 0 0 I
## 4 6102_18_9 6102 18 <NA> 0 0 I
## 5 6102_18_9 6102 18 <NA> 0 0 I
## 6 6102_439_26 6102 439 cold 17 0 I
## RNA_ng.ul total.yield_ng elution_vol_ul org_sample_ul target sq_all.runs_mean
## 1 19.90 258.70 15 70 Hemato 42.225
## 2 12.30 159.90 15 70 Hemato 42.225
## 3 34.70 451.10 15 70 Hemato 42.225
## 4 4.46 57.98 15 70 Hemato 32.544
## 5 4.46 57.98 15 70 Hemato 32.544
## 6 21.60 280.80 15 70 Hemato 32.544
## sample_pos.neg Library_ID
## 1 pos 329775
## 2 pos 329777
## 3 pos NA
## 4 neg NA
## 5 pos NA
## 6 neg 304428
Want to get average sqallmeans for each FRP…
avQpcr <- group_by(qpcr, FRP) %>%
summarize(avg_sq = mean(sq_all.runs_mean, na.rm=TRUE), trtmnt_tank, infection_status, maturity, sample_pos.neg) %>%
distinct_all() %>%
filter(trtmnt_tank == "cold" | trtmnt_tank == "warm" | trtmnt_tank == "ambient")
## `summarise()` has grouped output by 'FRP'. You can override using the `.groups` argument.
avQpcr
## # A tibble: 210 x 6
## # Groups: FRP [172]
## FRP avg_sq trtmnt_tank infection_status maturity sample_pos.neg
## <int> <dbl> <chr> <int> <chr> <chr>
## 1 6101 42.2 cold 0 I pos
## 2 6102 32.5 cold 0 I neg
## 3 6102 32.5 cold 0 I pos
## 4 6103 183. cold 1 I pos
## 5 6104 18.6 cold 0 I neg
## 6 6104 18.6 cold 0 I pos
## 7 6106 78.8 cold 0 M pos
## 8 6107 61.2 ambient 0 M neg
## 9 6107 61.2 ambient 0 M pos
## 10 6108 71.1 cold 1 I pos
## # … with 200 more rows
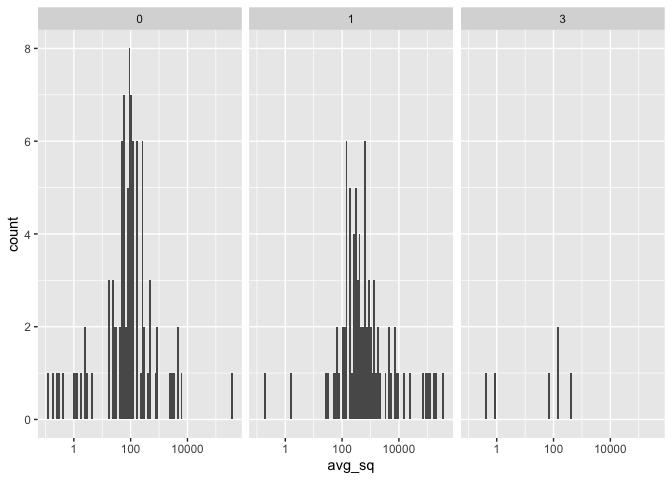
library(ggplot2)
ggplot(avQpcr, aes(x = avg_sq)) +
geom_histogram(bins = 100) +
scale_x_log10() +
facet_grid(~infection_status)
## Warning: Transformation introduced infinite values in continuous x-axis
## Warning: Removed 26 rows containing non-finite values (stat_bin).
Written on June 10, 2021
