Olympia Oyster Genome Read Check
Reposted from the FISH546 Project
I inadvertently started with ducks but now have rebooted with the Olympia oyster genome. The first step will be just seeing what we have..
What we expected
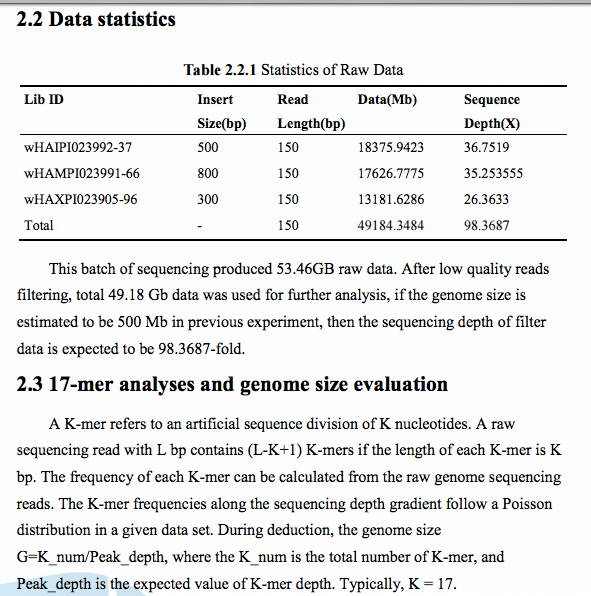
Received: 20160127
Format: FASTQ (Illumina HiSeq4000)
Location: http://owl.fish.washington.edu/nightingales/O_lurida/
Files:
| FILENAME | READS | INSERT SIZE | SEQUENCING |
|---|---|---|---|
| 151114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_1.fq.gz | 61253141 | 500bp | PE150 |
| 151114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_2.fq.gz | 61253141 | 500bp | PE150 |
| 151114_I191_FCH3Y35BCXX_L2_wHAMPI023991-66_1.fq.gz | 58755925 | 800bp | PE150 |
| 151114_I191_FCH3Y35BCXX_L2_wHAMPI023991-66_2.fq.gz | 53307837 | 800bp | PE150 |
| 151118_I137_FCH3KNJBBXX_L5_wHAXPI023905-96_1.fq.gz | 27731148 | 300bp | PE150 |
| 151118_I137_FCH3KNJBBXX_L5_wHAXPI023905-96_2.fq.gz | 43938762 | 300bp | PE150 |
| 160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCABDLAAPEI-62_1.fq.gz | 87198584 | 5000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCABDLAAPEI-62_2.fq.gz | 87198584 | 5000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCACDTAAPEI-75_1.fq.gz | 43766527 | 10000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCACDTAAPEI-75_2.fq.gz | 43766527 | 10000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCABDLAAPEI-62_1.fq.gz | 87135961 | 5000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCABDLAAPEI-62_2.fq.gz | 87135961 | 5000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCACDTAAPEI-75_1.fq.gz | 43138844 | 10000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCACDTAAPEI-75_2.fq.gz | 43138844 | 10000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L5_WHOSTibkDCAADWAAPEI-74_1.fq.gz | 95270180 | 2000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L5_WHOSTibkDCAADWAAPEI-74_2.fq.gz | 95270180 | 2000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L6_WHOSTibkDCAADWAAPEI-74_1.fq.gz | 92524416 | 2000bp | PE50 |
| 160103_I137_FCH3V5YBBXX_L6_WHOSTibkDCAADWAAPEI-74_2.fq.gz | 92524416 | 2000bp | PE50 |
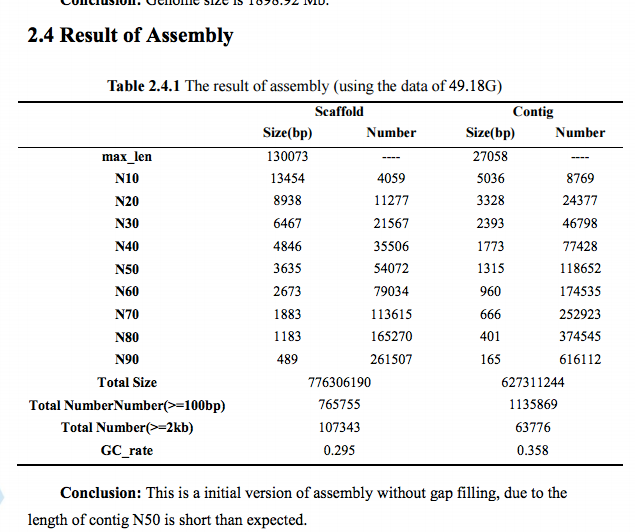
Data: Initial genome assembly
Received: 20160314
Formats: TXT, FASTA
Location: http://owl.fish.washington.edu/O_lurida_genome_assemblies_BGI/20160314/
Files:
| FILENAME | DESCRIPTION |
|---|---|
| md5.txt | Checksum file to verify integrity of files after downloading. |
| N50.txt | Contains limited stats on scaffolds provided. |
| scaffold.fa.fill | A FASTA file of scaffolds. |
max_length: 13711
N90 118 11683060
N80 127 9895195
N70 137 8233707
N60 150 6700118
N50 150 5237787
N40 150 3775455
N30 150 2313124
N20 209 964091
N10 494 234061
Total length: 2193496679
number>100bp 13627636
number>2000bp 13480
Data: Initial small insert library genome assembly
Received: 20160512
Formats: TXT, FASTA, PDF, PNG, FASTQ
Location: http://owl.fish.washington.edu/O_lurida_genome_assemblies_BGI/20160512/
Files:
FASTQ Files
| FILENAME | READS |
|---|---|
| 51114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_1.fq.gz.clean.dup.clean.gz | 61253141 |
| 151114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_2.fq.gz.clean.dup.clean.gz | 61253141 |
| 151114_I191_FCH3Y35BCXX_L2_wHAMPI023991-66_1.fq.gz.clean.dup.clean.gz | 58755925 |
| 151114_I191_FCH3Y35BCXX_L2_wHAMPI023991-66_2.fq.gz.clean.dup.clean.gz | 58755925 |
| 151118_I137_FCH3KNJBBXX_L5_wHAXPI023905-96_1.fq.gz.clean.dup.clean.gz | 43938762 |
| 151118_I137_FCH3KNJBBXX_L5_wHAXPI023905-96_2.fq.gz.clean.dup.clean.gz | 43938762 |
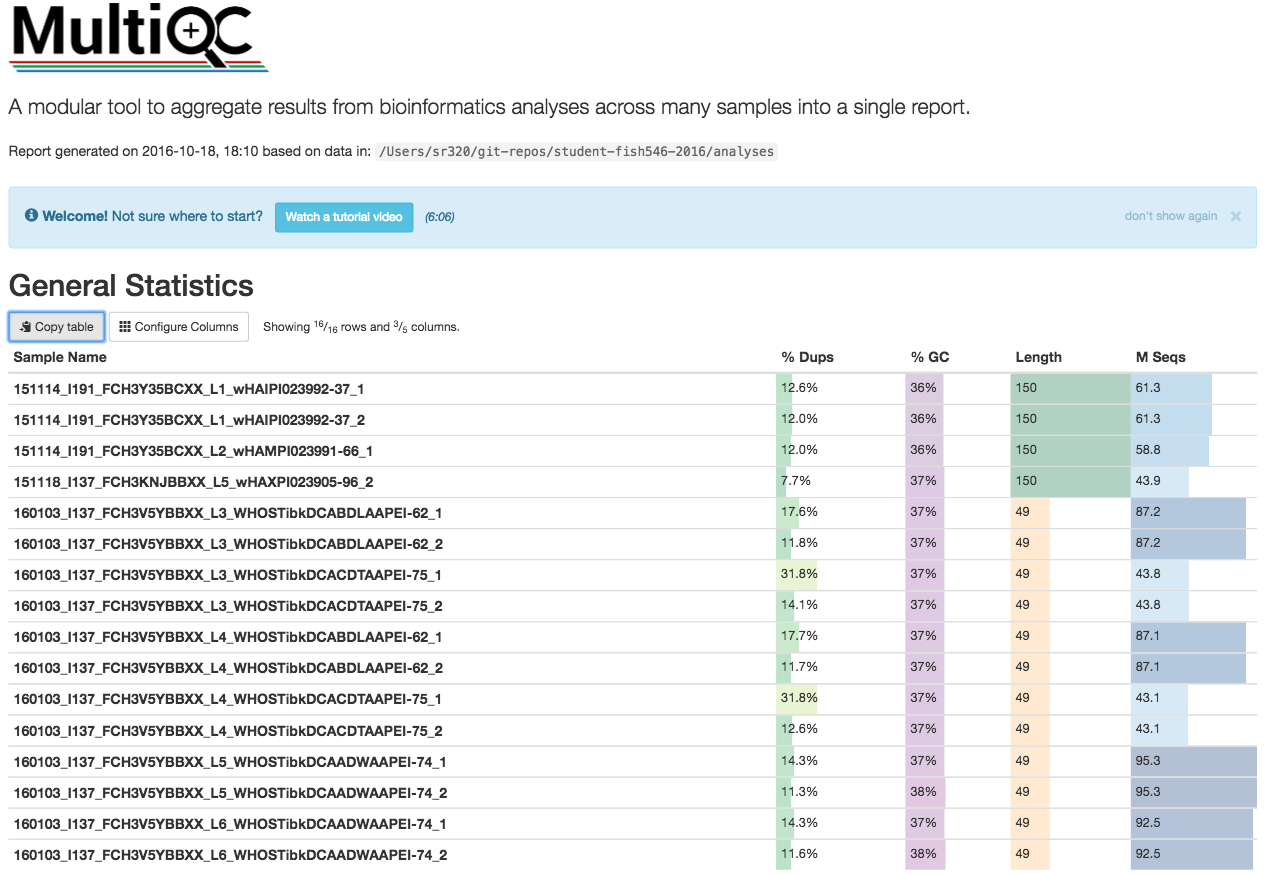
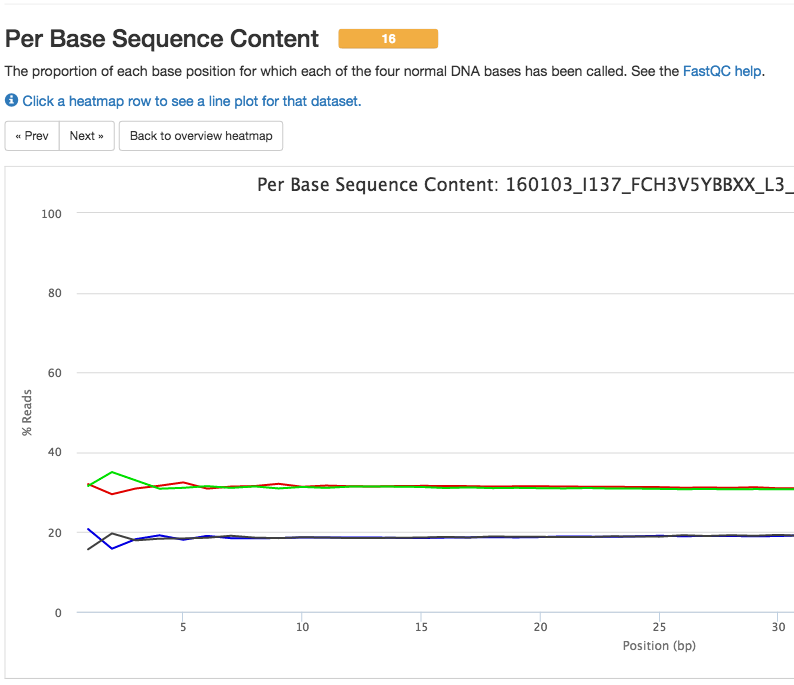
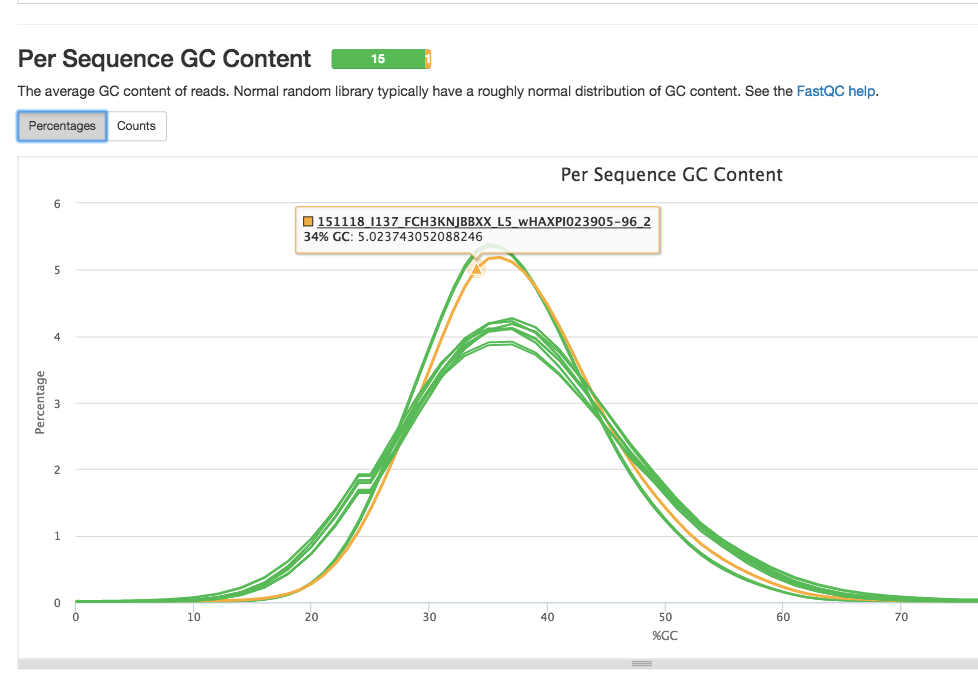
May 2016 Report
FastQC
$ ls nightingales/O_lurida/*I1*
nightingales/O_lurida/151114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_1.fq.gz*
nightingales/O_lurida/151114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_2.fq.gz*
nightingales/O_lurida/151114_I191_FCH3Y35BCXX_L2_wHAMPI023991-66_1.fq.gz*
nightingales/O_lurida/151114_I191_FCH3Y35BCXX_L2_wHAMPI023991-66_2.fq.gz*
nightingales/O_lurida/151118_I137_FCH3KNJBBXX_L5_wHAXPI023905-96_1.fq.gz*
nightingales/O_lurida/151118_I137_FCH3KNJBBXX_L5_wHAXPI023905-96_2.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCABDLAAPEI-62_1.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCABDLAAPEI-62_2.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCACDTAAPEI-75_1.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCACDTAAPEI-75_2.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCABDLAAPEI-62_1.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCABDLAAPEI-62_2.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCACDTAAPEI-75_1.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCACDTAAPEI-75_2.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L5_WHOSTibkDCAADWAAPEI-74_1.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L5_WHOSTibkDCAADWAAPEI-74_2.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L6_WHOSTibkDCAADWAAPEI-74_1.fq.gz*
nightingales/O_lurida/160103_I137_FCH3V5YBBXX_L6_WHOSTibkDCAADWAAPEI-74_2.fq.gz*
!/Applications/bioinfo/FastQC/fastqc \
nightingales/O_lurida/*I1* \
-t 6 \
-o /Users/sr320/git-repos/student-fish546-2016/analyses/
!ls /Users/sr320/git-repos/student-fish546-2016/analyses/
151114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_1.fq_fastqc.html
151114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_1.fq_fastqc.zip
151114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_2.fq_fastqc.html
151114_I191_FCH3Y35BCXX_L1_wHAIPI023992-37_2.fq_fastqc.zip
151114_I191_FCH3Y35BCXX_L2_wHAMPI023991-66_1.fq_fastqc.html
151114_I191_FCH3Y35BCXX_L2_wHAMPI023991-66_1.fq_fastqc.zip
151118_I137_FCH3KNJBBXX_L5_wHAXPI023905-96_2.fq_fastqc.html
151118_I137_FCH3KNJBBXX_L5_wHAXPI023905-96_2.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCABDLAAPEI-62_1.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCABDLAAPEI-62_1.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCABDLAAPEI-62_2.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCABDLAAPEI-62_2.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCACDTAAPEI-75_1.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCACDTAAPEI-75_1.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCACDTAAPEI-75_2.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L3_WHOSTibkDCACDTAAPEI-75_2.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCABDLAAPEI-62_1.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCABDLAAPEI-62_1.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCABDLAAPEI-62_2.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCABDLAAPEI-62_2.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCACDTAAPEI-75_1.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCACDTAAPEI-75_1.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCACDTAAPEI-75_2.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L4_WHOSTibkDCACDTAAPEI-75_2.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L5_WHOSTibkDCAADWAAPEI-74_1.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L5_WHOSTibkDCAADWAAPEI-74_1.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L5_WHOSTibkDCAADWAAPEI-74_2.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L5_WHOSTibkDCAADWAAPEI-74_2.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L6_WHOSTibkDCAADWAAPEI-74_1.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L6_WHOSTibkDCAADWAAPEI-74_1.fq_fastqc.zip
160103_I137_FCH3V5YBBXX_L6_WHOSTibkDCAADWAAPEI-74_2.fq_fastqc.html
160103_I137_FCH3V5YBBXX_L6_WHOSTibkDCAADWAAPEI-74_2.fq_fastqc.zip
Seemed to have missed a few..
66,96
MultiQC
Full Report:
http://owl.fish.washington.edu/halfshell/working-directory/16-10-19/multiqc_report.html#
!multiqc /Users/sr320/git-repos/student-fish546-2016/analyses