Refreshing the Caligus
Updated: February 05
Trimmed reads again increased mapping efficiency
Mapping efficiency: 28.2%
Mapping efficiency: 27.9%
Mapping efficiency: 21.8%
Mapping efficiency: 21.5%
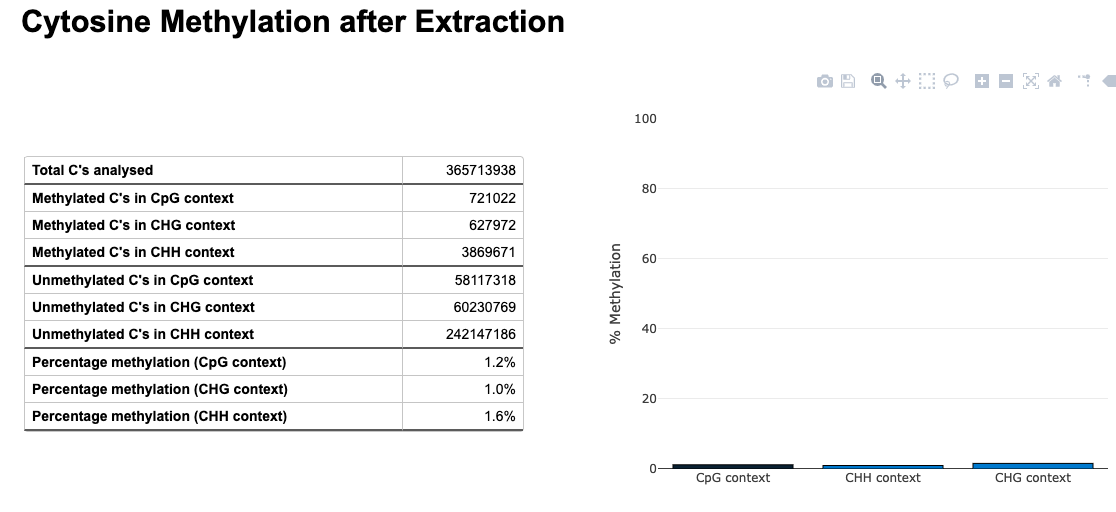
Seems ok.. now will “post-process”. Specifically https://d.pr/n/hwSCRy
And check out 5x coverage tracks!

output directory: https://gannet.fish.washington.edu/seashell/bu-mox/scrubbed/020320-cal/
Updated: February 03
Mapping efficiency did in fact almost double
Mapping efficiency: 15.9%
Mapping efficiency: 15.6%
Mapping efficiency: 11.6%
Mapping efficiency: 11.4%
Average methylation about 2%

Will now try trimmed reads https://d.pr/n/JnNCn9
Updated: January 30
With very low mapping efficiency (~8) https://github.com/RobertsLab/resources/issues/821
I reran https://d.pr/n/z8SSi3
with a few alignment tweaks
--non_directional \
--dovetail \
I expect the non_directional (presuming accurate) could double efficiency.
2019-12-31
In an effort to back into lice, I tried rerunning bismark such that we could visualize with reasonable thresholds. job: https://d.pr/n/lyMYD9
Unfortunately this is all that was created
[sr320@mox1 1219]$ ls
merged_CpG_evidence.cov_10x.bedgraph Sealice_F1_S20_R2_001_trimmed.5bp_3prime.fq.gz_G_to_A.fastq slurm-1580450.out
*merged_CpG_evidence.cov_5x.bedgraph Sealice_F2_S22_R1_001_trimmed.5bp_3prime.fq.gz_C_to_T.fastq slurm-1580615.out
Sealice_F1_S20_R1_001_trimmed.5bp_3prime.fq.gz_C_to_T.fastq Sealice_F2_S22_R2_001_trimmed.5bp_3prime.fq.gz_G_to_A.fastq slurm-1590565.out
Did see one issue
[FATAL ERROR]: Number of bisulfite transformed reads are not equal between Read 1 (#226106788) and Read 2 (#225857063).
Possible causes: file truncation, or as a result of specifying read pairs that do not belong to each other?! Please re-specify file names! Exiting...
Going to run raw reads.
dc277e523adb9a09b0fe139432cc0fc3 Sealice_F1_S20_L001_R1_001.fastq.gz
2e42b2d6f9d2cf8d610001921426f416 Sealice_F1_S20_L001_R2_001.fastq.gz
deacebe41dc6826091571aebd235fe1a Sealice_F1_S20_L002_R1_001.fastq.gz
d838e9fd5e14d2fb3d3629c9d4af2ddc Sealice_F1_S20_L002_R2_001.fastq.gz
2acb16a5f192e42a9cc057ba962a9950 Sealice_F2_S22_L001_R1_001.fastq.gz
5812d8a1d8e0af717743612d88022fd0 Sealice_F2_S22_L001_R2_001.fastq.gz
6082314c6680524041b7b99a79f7e913 Sealice_F2_S22_L002_R1_001.fastq.gz
0c944326b09da6f7d2a186505b749022 Sealice_F2_S22_L002_R2_001.fastq.gz
Here is the new job script: https://d.pr/n/qleuJ3
