Reproducing whole-Duck analyses, with a few bumps
In preparation for the soon submission of the geoduck genome paper, I re-ran some mox jobs to see if we could reproduce the paper, particular with regard to the new naming scheme. The two scripts were 1217_1300.sh and 1218_1000.sh.
“New” aspects of both include coverage2cytosine
find *deduplicated.bismark.cov.gz \
| xargs basename -s _R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz \
| xargs -I{} ${bismark_dir}/coverage2cytosine \
--genome_folder ${genome_folder} \
-o {} \
--merge_CpG \
--zero_based \
{}_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
and for the 1218_1000 took it a step further and created bedgraphs
for f in *merged_CpG_evidence.cov
do
STEM=$(basename "${f}" .CpG_report.merged_CpG_evidence.cov)
cat "${f}" | awk '{if ($5+$6 >= 5) {print $1, $2, $3, $4}}' \
> "${STEM}"_5x.bedgraph
done
Did it work?
Well output from each job can be found here and here.
At least for the latter there seems to be an issue.
Stored sequence information of 18 chromosomes/scaffolds in total
==============================================================================
Methylation information will now be written into a genome-wide cytosine report
==============================================================================
>>> Writing genome-wide cytosine report to: EPI-103_S27_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.bismark.cov.gz.CpG_report.txt <<<
gzip: EPI-103_S27_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.bismark.cov.gz_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz: No such file or directory
No last chromosome was defined, something must have gone wrong while reading the data in (e.g. specified wrong file path for a gzipped coverage file?). Please check your command!
xargs: /gscratch/srlab/programs/Bismark-0.21.0/coverage2cytosine: exited with status 255; aborting
cat: merged_CpG_evidence.cov: No such file or directory
On the other hand there seems to no issue with 1217
>>> Writing genome-wide cytosine report to: EPI-44_S41_L005.CpG_report.txt <<<
Storing all covered cytosine positions for chromosome: Scaffold_01
Writing cytosine report for chromosome Scaffold_01 (stored 588975 different covered positions)
Writing cytosine report for chromosome Scaffold_02 (stored 453991 different covered positions)
Writing cytosine report for chromosome Scaffold_03 (stored 365994 different covered positions)
Writing cytosine report for chromosome Scaffold_04 (stored 416373 different covered positions)
Writing cytosine report for chromosome Scaffold_05 (stored 373321 different covered positions)
Writing cytosine report for chromosome Scaffold_06 (stored 363497 different covered positions)
Writing cytosine report for chromosome Scaffold_07 (stored 273122 different covered positions)
Writing cytosine report for chromosome Scaffold_08 (stored 377813 different covered positions)
Writing cytosine report for chromosome Scaffold_09 (stored 236744 different covered positions)
Writing cytosine report for chromosome Scaffold_10 (stored 316503 different covered positions)
Writing cytosine report for chromosome Scaffold_11 (stored 305652 different covered positions)
Writing cytosine report for chromosome Scaffold_12 (stored 327312 different covered positions)
Writing cytosine report for chromosome Scaffold_13 (stored 268339 different covered positions)
Writing cytosine report for chromosome Scaffold_14 (stored 341289 different covered positions)
Writing cytosine report for chromosome Scaffold_15 (stored 267283 different covered positions)
Writing cytosine report for chromosome Scaffold_16 (stored 210410 different covered positions)
Writing cytosine report for chromosome Scaffold_17 (stored 229577 different covered positions)
Writing cytosine report for last chromosome Scaffold_18 (stored 164350 different covered positions)
Finished writing out cytosine report for covered chromosomes (processed 18 chromosomes/scaffolds in total)
Now processing chromosomes that were not covered by any methylation calls in the coverage file...
All chromosomes in the genome were covered by at least some reads. coverage2cytosine processing complete.
Now merging top and bottom strand CpGs into a single CG dinucleotide entity (reading from file >>EPI-44_S41_L005.CpG_report.txt<<, in output directory '')
>>> Writing a new coverage file with top and bottom strand CpG methylation evidence merged to EPI-44_S41_L005.CpG_report.merged_CpG_evidence.cov <<<
CpG merging complete. Good luck finding DMRs with bsseq!
1218 did create more Files
[sr320@mox1 1217]$ ls | wc -l
991
[sr320@mox1 1218]$ ls | wc -l
1758
Will check against bedgraph files created in Jupyter https://github.com/sr320/nb-2019/blob/master/P_generosa/40-cov2cyt-zero.ipynb
The bedgraphs are @: https://gannet.fish.washington.edu/seashell/bu-serine-wd/19-12-17/zero-based/bg/
and I also needed generate files with count data and those files can be found @: https://gannet.fish.washington.edu/seashell/bu-serine-wd/19-12-17/zero-based/glsm/
Interesting it would seem 1218 somehow hit the dedup/deddup/dup issue.
[sr320@mox1 1217]$ ls *-41_*
CHG_OB_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CHG_OT_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CHH_OB_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CHH_OT_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CpG_OB_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CpG_OT_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
EPI-41_S38_L005.CpG_report.merged_CpG_evidence.cov
EPI-41_S38_L005.CpG_report.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.bam
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted.bam
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted.bam.bai
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_PE_report.html
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_PE_report.txt
[sr320@mox1 1218]$ ls *-41_*
CHG_OB_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.txt
CHG_OB_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CHG_OT_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.txt
CHG_OT_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CHH_OB_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.txt
CHH_OB_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CHH_OT_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.txt
CHH_OT_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CpG_OB_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.txt
CpG_OB_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
CpG_OT_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.txt
CpG_OT_EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
EPI-41_S38_L005_5x.bedgraph
EPI-41_S38_L005.CpG_report.merged_CpG_evidence.cov
EPI-41_S38_L005.CpG_report.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.bam
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.bam
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.bedGraph.gz
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.bismark.cov.gz
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.M-bias.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.sorted.bam
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated.sorted.bam.bai
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplicated_splitting_report.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.deduplication_report.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted.bam
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted.bam.bai
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_PE_report.html
EPI-41_S38_L005_R1_001_val_1_bismark_bt2_PE_report.txt
This is quite hard to explain …. given no real differences.
[sr320@mox1 2019]$ diff 1217_1300.sh 1218_1000.sh
3c3
< #SBATCH --job-name=re-duck
---
> #SBATCH --job-name=re-d2
5,6c5,6
< #SBATCH --account=srlab
< #SBATCH --partition=srlab
---
> #SBATCH --account=coenv
> #SBATCH --partition=coenv
11c11
< #SBATCH --time=30-00:00:00
---
> #SBATCH --time=6-00:00:00
17c17
< #SBATCH --chdir=/gscratch/scrubbed/sr320/1217/
---
> #SBATCH --chdir=/gscratch/scrubbed/sr320/1218/
31,36c31,36
<
< ${bismark_dir}/bismark_genome_preparation \
< --verbose \
< --parallel 28 \
< --path_to_aligner ${bowtie2_dir} \
< ${genome_folder}
---
> #
> #${bismark_dir}/bismark_genome_preparation \
> #--verbose \
> #--parallel 28 \
> #--path_to_aligner ${bowtie2_dir} \
> #${genome_folder}
42c42
< -genome /gscratch/srlab/sr320/data/geoduck/v01 \
---
> -genome ${genome_folder} \
107a108,122
> for f in merged_CpG_evidence.cov
> do
> STEM=$(basename "${f}" .CpG_report.merged_CpG_evidence.cov)
> cat "${f}" | awk '{if ($5+$6 >= 10) {print $1, $2, $3, $4}}' \
> > "${STEM}"_10x.bedgraph
> done
>
>
>
> for f in *merged_CpG_evidence.cov
> do
> STEM=$(basename "${f}" .CpG_report.merged_CpG_evidence.cov)
> cat "${f}" | awk '{if ($5+$6 >= 5) {print $1, $2, $3, $4}}' \
> > "${STEM}"_5x.bedgraph
> done
\ No newline at end of file
Different issue - found missing * for 10x bedGraph
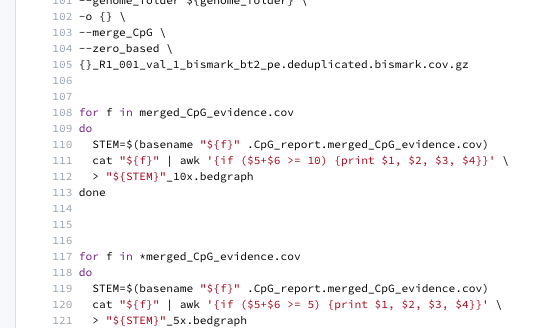
Will go ahead and run again fixing the *, crossing fingers on dedup/dedup issue, and adding code to get tab file with raw count data post merging.
New job: https://d.pr/n/ZbsU6s
Update
output: https://gannet.fish.washington.edu/seashell/bu-mox/scrubbed/1231/
