Where are the SNPs?
1 Background
https://github.com/sr320/ceabigr/issues/78
At minimum, identifying C->T SNPs on both strands so we have a list of SNPs from the EpiDiverse output.
Other tasks that build upon that:
- Compare Bs-snper and EpiDiverse SNP lists. Determine which source our final C->T SNP list should come from
- Remove SNPs from methylKit analysis
- Remove SNPs and recalculate average gene methylation and CV of gene methylation for modeling analyses
There are two ways we have used to get genetic information from DNA methylation data. - EPIDiverse - BS-SNPer
2 Epidiverse
Sam ran Epidiverse
https://github.com/sr320/ceabigr/issues/69#issuecomment-1258238481
- Notebook
- VCF Directory - https://gannet.fish.washington.edu/Atumefaciens/20220921-cvir-ceabigr-nextflow-epidiverse-snp/snps/vcf/
- results dir - https://gannet.fish.washington.edu/Atumefaciens/20220921-cvir-ceabigr-nextflow-epidiverse-snp/snps/stats/
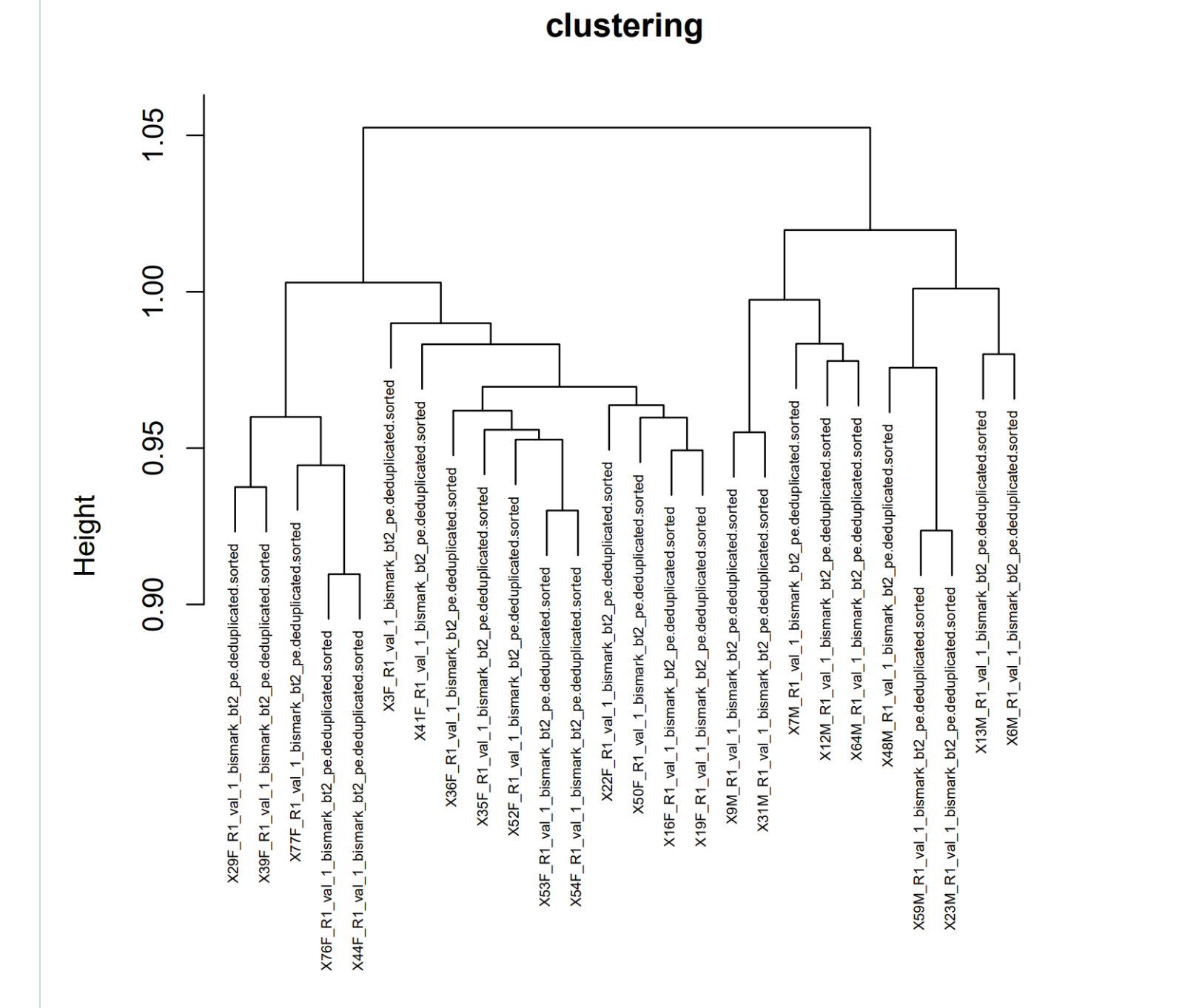
Next step for capturing SNP info in Epidiverse Workflow is merging.
3 BS-SNPer
3.1 draft paper
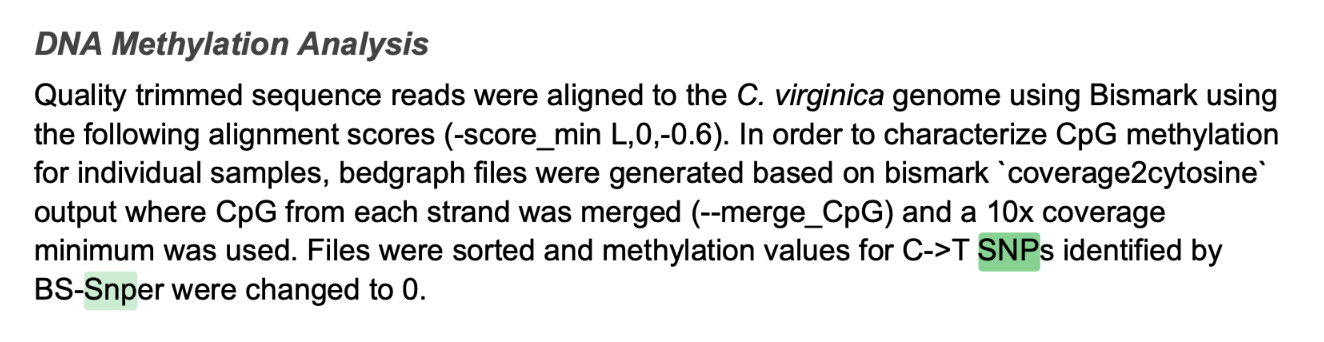
3.2 Wiki
10x coverage files (SNP corrected)
at https://gannet.fish.washington.edu/seashell/ceabigr/output/methylation-landscape/
\(sampleID\)_R1_val_1_10x.SNPcorr.bedgraph
3.3 issue (backtracking)
see https://github.com/sr320/ceabigr/issues/10
# Directories and programs
bismark_dir="/gscratch/srlab/programs/Bismark-0.22.3/"
bowtie2_dir="/gscratch/srlab/programs/bowtie2-2.3.4.1-linux-x86_64/"
samtools="/gscratch/srlab/programs/samtools-1.9/samtools/"
reads_dir="/gscratch/srlab/sr320/data/cvirg/"
fastqc="/gscratch/srlab/programs/fastqc_v0.11.9/fastqc"
genome_folder="/gscratch/srlab/sr320/data/Cvirg-genome/"
bcftools_dir="/gscratch/srlab/programs/bcftools-1.9/"
htlslib_dir="/gscratch/srlab/programs/htslib-1.9/"
source /gscratch/srlab/programs/scripts/paths.sh
#
# Ah sorry, I always forget about this with bcftools.
#
#Loop through files and compress like this:
#
# bcftools view -Oz -o compressed.vcf.gz plain.vcf
# htsfile compressed.vcf.gz
# bcftools index compressed.vcf.gz
# FILES=$(ls *vcf)
#
# for file in ${FILES}
# do
# NAME=$(echo ${file} | awk -F "." '{print $1}')
# echo ${NAME}
#
# /gscratch/srlab/programs/bcftools-1.9/bcftools view -O z -o ${NAME}.compressed.vcf.gz \
# ${NAME}.SNP-results.vcf
# /gscratch/srlab/programs/htslib-1.9/htsfile ${NAME}.compressed.vcf.gz
# /gscratch/srlab/programs/bcftools-1.9/bcftools index ${NAME}.compressed.vcf.gz
# done
#
#
# /gscratch/srlab/programs/bcftools-1.9/bcftools merge \
# 12M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 44F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 13M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 48M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 16F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 50F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 19F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 52F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 22F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 53F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 23M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 54F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 29F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 59M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 31M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 64M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 35F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 6M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 36F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 76F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 39F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 77F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 3F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 7M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 41F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 9M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# --merge all \
# --threads 20 \
# -O v \
# -o Cv_10x_merged.vcf
/gscratch/srlab/programs/vcftools-0.1.16/bin/vcftools \
--vcf Cv_10x_merged.vcf \
--recode --recode-INFO-all \
--min-alleles 2 --max-alleles 2 \
--max-missing 0.5 \
--mac 2 \
--out Cv_10x_merged.filtered
Merged VCF - 16G https://gannet.fish.washington.edu/seashell/bu-mox/scrubbed/021722-vcfmerge/Cv_10x_merged.vcf
filtered file.. https://github.com/sr320/ceabigr/blob/main/data/Cv_10x_fxmerge_05.recode.vcf
head ../data/Cv_10x_fxmerge_05.recode.vcf
## ##fileformat=VCFv4.2
## ##FILTER=<ID=PASS,Description="All filters passed">
## ##fileDate= 20220213
## ##bssnperVersion=1.1
## ##bssnperCommand=--fa ../data/Cvirginica_v300.fa --input /home/sr320/github/2018_L18-adult-methylation/bg_data/12M_R1_val_1_bismark_bt2_pe.deduplicated.sorted.bam --output ../analyses/bsnp02/12M_R1_val_1_bismark_bt2_pe.SNP-candidates.txt --methcg ../analyses/bsnp02/12M_R1_val_1_bismark_bt2_pe.CpG-meth-info.tab --methchg ../analyses/bsnp02/12M_R1_val_1_bismark_bt2_pe.CHG-meth-info.tab --methchh ../analyses/bsnp02/12M_R1_val_1_bismark_bt2_pe.CHH-meth-info.tab --minhetfreq 0.1 --minhomfreq 0.85 --minquali 15 --mincover 10 --maxcover 1000 --minread2 2 --errorate 0.02 --mapvalue 20
## ##reference=file://../data/Cvirginica_v300.fa
## ##Bisulfite=directional>
## ##contig=<ID=NC_035780.1,length=65668440>
## ##contig=<ID=NC_035781.1,length=61752955>
## ##contig=<ID=NC_035782.1,length=77061148>
tail -2 ../data/Cv_10x_fxmerge_05.recode.vcf
## NC_007175.2 12774 . C T 1000 PASS DP=91497;ADF=0,0;ADR=0,220;AD=0,220;AN=52;AC=26 GT:DP:ADF:ADR:AD:BSD:BSQ:ALFR 0/1:407:0,0:0,407:0,407:0,0,223,0,0,407,0,0:0,0,36,0,0,37,0,0:0,1 0/1:5786:0,0:8,5778:8,5778:0,0,4267,1,3,5778,8,0:0,0,36,25,37,36,36,0:0.001,0.999 0/1:557:0,0:1,556:1,556:0,0,391,0,1,556,1,0:0,0,36,0,37,37,37,0:0.002,0.998 0/1:819:0,0:0,819:0,819:0,0,695,0,0,819,0,0:0,0,36,0,0,36,0,0:0,1 0/1:2838:0,0:6,2832:6,2832:0,0,1829,0,1,2832,6,0:0,0,36,0,37,36,35,0:0.002,0.998 0/1:4256:0,0:9,4247:9,4247:0,0,2781,0,4,4247,9,1:0,0,36,0,34,36,37,37:0.002,0.998 0/1:5216:0,0:9,5207:9,5207:0,0,6064,0,2,5207,9,1:0,0,36,0,31,36,37,37:0.002,0.998 0/1:3184:0,0:2,3182:2,3182:0,0,1748,0,1,3182,2,1:0,0,36,0,37,36,37,25:0.001,0.999 0/1:5953:0,0:7,5946:7,5946:0,0,3806,0,0,5946,7,1:0,0,36,0,0,37,35,25:0.001,0.999 0/1:7521:0,0:10,7511:10,7511:0,0,5104,5,7,7511,10,1:0,0,36,25,35,36,36,25:0.001,0.999 0/1:386:0,0:2,384:2,384:0,0,299,0,0,384,2,0:0,0,36,0,0,37,37,0:0.005,0.995 0/1:4995:0,0:8,4987:8,4987:0,0,3422,1,1,4987,8,1:0,0,36,25,37,36,37,37:0.002,0.998 0/1:2953:0,0:2,2951:2,2951:0,0,2036,0,1,2951,2,0:0,0,36,0,37,36,37,0:0.001,0.999 0/1:288:0,0:2,286:2,286:0,0,167,0,0,286,2,0:0,0,36,0,0,36,37,0:0.007,0.993 0/1:283:0,0:4,279:4,279:0,0,195,0,0,279,4,0:0,0,36,0,0,36,37,0:0.014,0.986 0/1:747:0,0:3,744:3,744:0,0,554,0,0,744,3,0:0,0,36,0,0,37,37,0:0.004,0.996 0/1:8439:0,0:17,8422:17,8422:0,0,6163,3,5,8422,17,0:0,0,36,37,37,36,36,0:0.002,0.998 0/1:597:0,0:0,597:0,597:0,0,494,2,3,597,0,0:0,0,36,25,37,36,0,0:0,1 0/1:6748:0,0:8,6740:8,6740:0,0,5233,1,6,6740,8,0:0,0,36,25,37,36,37,0:0.001,0.999 0/1:6916:0,0:8,6908:8,6908:0,0,6197,1,2,6908,8,0:0,0,36,25,37,37,37,0:0.001,0.999 0/1:7641:0,0:11,7630:11,7630:0,0,6446,2,5,7630,11,0:0,0,36,25,37,36,36,0:0.001,0.999 0/1:7115:0,0:4,7111:4,7111:1,0,6214,1,4,7111,4,0:25,0,36,25,37,36,37,0:0.001,0.999 0/1:5198:0,0:3,5195:3,5195:0,0,4280,0,1,5195,3,1:0,0,36,0,37,36,37,37:0.001,0.999 0/1:521:0,0:2,519:2,519:0,0,287,0,0,519,2,0:0,0,36,0,0,36,37,0:0.004,0.996 0/1:1913:0,0:4,1909:4,1909:0,0,1107,0,0,1909,4,2:0,0,36,0,0,37,37,37:0.002,0.998 0/1:220:0,0:0,220:0,220:0,0,158,0,0,220,0,0:0,0,36,0,0,37,0,0:0,1
## NC_007175.2 15247 . C T 1000 PASS DP=43802;ADF=0,0;ADR=0,57;AD=0,57;AN=52;AC=26 GT:DP:ADF:ADR:AD:BSD:BSQ:ALFR 0/1:112:0,0:0,112:0,112:0,0,322,0,0,112,0,0:0,0,36,0,0,36,0,0:0,1 0/1:2287:0,0:5,2282:5,2282:0,0,3893,1,2,2282,5,1:0,0,36,25,37,36,35,25:0.002,0.998 0/1:174:0,0:0,174:0,174:0,0,408,0,0,174,0,0:0,0,37,0,0,36,0,0:0,1 0/1:312:0,0:0,312:0,312:0,0,805,0,0,312,0,0:0,0,36,0,0,36,0,0:0,1 0/1:1310:0,0:0,1310:0,1310:0,0,1883,0,0,1310,0,0:0,0,36,0,0,36,0,0:0,1 0/1:2014:0,0:8,2006:8,2006:0,0,2965,0,3,2006,8,0:0,0,36,0,33,36,36,0:0.004,0.996 0/1:4017:0,0:7,4010:7,4010:0,0,6081,0,6,4010,7,0:0,0,36,0,37,36,37,0:0.002,0.998 0/1:1398:0,0:2,1396:2,1396:0,0,1817,0,2,1396,2,0:0,0,36,0,37,36,37,0:0.001,0.999 0/1:2263:0,0:2,2261:2,2261:0,0,3922,1,3,2261,2,0:0,0,36,25,33,36,37,0:0.001,0.999 0/1:3506:0,0:2,3504:2,3504:0,0,4504,0,3,3504,2,0:0,0,36,0,37,36,37,0:0.001,0.999 0/1:140:0,0:2,138:2,138:0,0,291,0,0,138,2,0:0,0,36,0,0,36,37,0:0.014,0.986 0/1:2187:0,0:0,2187:0,2187:0,0,3267,0,0,2187,0,1:0,0,36,0,0,36,0,25:0,1 0/1:1099:0,0:3,1096:3,1096:1,0,1999,0,0,1096,3,0:37,0,36,0,0,36,29,0:0.003,0.997 0/1:106:0,0:0,106:0,106:0,0,182,0,0,106,0,0:0,0,36,0,0,36,0,0:0,1 0/1:136:0,0:0,136:0,136:0,0,249,0,0,136,0,0:0,0,36,0,0,36,0,0:0,1 0/1:230:0,0:0,230:0,230:0,0,540,0,0,230,0,0:0,0,36,0,0,36,0,0:0,1 0/1:4059:0,0:9,4050:9,4050:0,0,5635,1,5,4050,9,2:0,0,36,37,37,36,36,31:0.002,0.998 0/1:188:0,0:0,188:0,188:0,0,624,1,0,188,0,0:0,0,36,25,0,36,0,0:0,1 0/1:3105:0,0:3,3102:3,3102:1,0,4920,0,4,3102,3,1:25,0,36,0,34,36,37,25:0.001,0.999 0/1:3066:0,0:3,3063:3,3063:0,0,6416,2,3,3063,3,0:0,0,36,37,33,36,37,0:0.001,0.999 0/1:4533:0,0:6,4527:6,4527:0,0,5452,1,2,4527,6,0:0,0,36,37,37,36,35,0:0.001,0.999 0/1:3451:0,0:5,3446:5,3446:0,0,6195,2,1,3446,5,2:0,0,36,25,37,36,37,37:0.001,0.999 0/1:2841:0,0:0,2841:0,2841:1,0,4298,0,3,2841,0,1:25,0,36,0,33,36,0,25:0,1 0/1:159:0,0:0,159:0,159:0,0,373,0,0,159,0,0:0,0,36,0,0,36,0,0:0,1 0/1:1052:0,0:4,1048:4,1048:0,0,1293,1,0,1048,4,0:0,0,36,25,0,36,37,0:0.004,0.996 0/1:57:0,0:0,57:0,57:0,0,165,0,0,57,0,0:0,0,37,0,0,35,0,0:0,1
