FROGER Day 03
We have some mapping done and will push forward with what we have his AM, which is a 100M subset of Mcap.
[sr320@mox2 030520-fr01]$ cat job11.sh
#!/bin/bash
## Job Name
#SBATCH --job-name=full-u10M
## Allocation Definition
#SBATCH --account=srlab
#SBATCH --partition=srlab
## Resources
## Nodes (We only get 1, so this is fixed)
#SBATCH --nodes=1
## Walltime (days-hours:minutes:seconds format)
#SBATCH --time=2-00:00:00
## Memory per node
#SBATCH --mem=100G
#SBATCH --mail-type=ALL
#SBATCH --mail-user=sr320@uw.edu
## Specify the working directory for this job
#SBATCH --chdir=/gscratch/scrubbed/sr320/030520-fr01/
# Directories and programs
bismark_dir="/gscratch/srlab/programs/Bismark-0.21.0"
bowtie2_dir="/gscratch/srlab/programs/bowtie2-2.3.4.1-linux-x86_64/"
samtools="/gscratch/srlab/programs/samtools-1.9/samtools"
source /gscratch/srlab/programs/scripts/paths.sh
# ${bismark_dir}/bismark_genome_preparation \
# --verbose \
# --parallel 28 \
# --path_to_aligner ${bowtie2_dir} \
# ${genome_folder}
reads_dir="/gscratch/srlab/sr320/data/froger/trim/Mc/"
genome_folder="/gscratch/srlab/sr320/data/froger/Mcap_Genome/"
find ${reads_dir}*2020*_R1_001.fastq.gz \
| xargs basename -s _R1_001.fastq.gz | xargs -I{} ${bismark_dir}/bismark \
--path_to_bowtie ${bowtie2_dir} \
-genome ${genome_folder} \
-p 4 \
-score_min L,0,-0.6 \
-u 10000000 \
--non_directional \
-1 ${reads_dir}{}_R1_001.fastq.gz \
-2 ${reads_dir}{}_R2_001.fastq.gz \
-o Mcap_full-u1M
reads_dir="/gscratch/srlab/sr320/data/froger/trim/Pa/"
genome_folder="/gscratch/srlab/sr320/data/froger/Pact_Genome/"
find ${reads_dir}*2020*_R1_001.fastq.gz \
| xargs basename -s _R1_001.fastq.gz | xargs -I{} ${bismark_dir}/bismark \
--path_to_bowtie ${bowtie2_dir} \
-genome ${genome_folder} \
-p 4 \
-score_min L,0,-0.6 \
-u 10000000 \
--non_directional \
-1 ${reads_dir}{}_R1_001.fastq.gz \
-2 ${reads_dir}{}_R2_001.fastq.gz \
-o Pact_full-u1M
reads_dir="/gscratch/srlab/sr320/data/froger/trim/Pa/"
genome_folder="/gscratch/srlab/sr320/data/froger/Pdam_Genome/"
find ${reads_dir}*2020*_R1_001.fastq.gz \
| xargs basename -s _R1_001.fastq.gz | xargs -I{} ${bismark_dir}/bismark \
--path_to_bowtie ${bowtie2_dir} \
-genome ${genome_folder} \
-p 4 \
-score_min L,0,-0.6 \
-u 10000000 \
--non_directional \
-1 ${reads_dir}{}_R1_001.fastq.gz \
-2 ${reads_dir}{}_R2_001.fastq.gz \
-o Pdam_full-u1M
reads_dir="/gscratch/scrubbed/samwhite/outputs/20200305_methcompare_fastp_trimming/"
genome_folder="/gscratch/srlab/sr320/data/lambda/"
find ${reads_dir}*2020*_R1_001.fastq.gz \
| xargs basename -s _R1_001.fastq.gz | xargs -I{} ${bismark_dir}/bismark \
--path_to_bowtie ${bowtie2_dir} \
-genome ${genome_folder} \
-p 4 \
-score_min L,0,-0.6 \
-u 10000000 \
--non_directional \
-1 ${reads_dir}{}_R1_001.fastq.gz \
-2 ${reads_dir}{}_R2_001.fastq.gz \
-o lambda_full-u1M
[sr320@mox2 030520-fr01]$ ls -lh Mcap_full-u1M/
total 7.6G
-rw-r--r-- 1 sr320 hyak-coenv 898M Mar 7 01:19 Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.bam
-rw-r--r-- 1 sr320 hyak-coenv 1.9K Mar 7 01:19 Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_PE_report.txt
-rw-r--r-- 1 sr320 hyak-coenv 931M Mar 7 01:55 Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.bam
-rw-r--r-- 1 sr320 hyak-coenv 1.9K Mar 7 01:55 Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_PE_report.txt
-rw-r--r-- 1 sr320 hyak-coenv 930M Mar 7 02:31 Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.bam
-rw-r--r-- 1 sr320 hyak-coenv 1.9K Mar 7 02:31 Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_PE_report.txt
-rw-r--r-- 1 sr320 hyak-coenv 1.2G Mar 7 03:16 Meth13.fastp-trim.202003062040_R1_001_bismark_bt2_pe.bam
-rw-r--r-- 1 sr320 hyak-coenv 1.9K Mar 7 03:16 Meth13.fastp-trim.202003062040_R1_001_bismark_bt2_PE_report.txt
-rw-r--r-- 1 sr320 hyak-coenv 1.1G Mar 7 03:59 Meth14.fastp-trim.202003064415_R1_001_bismark_bt2_pe.bam
-rw-r--r-- 1 sr320 hyak-coenv 1.9K Mar 7 03:59 Meth14.fastp-trim.202003064415_R1_001_bismark_bt2_PE_report.txt
-rw-r--r-- 1 sr320 hyak-coenv 1.1G Mar 7 04:41 Meth15.fastp-trim.202003065503_R1_001_bismark_bt2_pe.bam
-rw-r--r-- 1 sr320 hyak-coenv 1.9K Mar 7 04:41 Meth15.fastp-trim.202003065503_R1_001_bismark_bt2_PE_report.txt
-rw-r--r-- 1 sr320 hyak-coenv 556M Mar 7 05:06 Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.bam
-rw-r--r-- 1 sr320 hyak-coenv 1.9K Mar 7 05:06 Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_PE_report.txt
-rw-r--r-- 1 sr320 hyak-coenv 527M Mar 7 05:30 Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.bam
-rw-r--r-- 1 sr320 hyak-coenv 1.9K Mar 7 05:30 Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_PE_report.txt
-rw-r--r-- 1 sr320 hyak-coenv 516M Mar 7 05:54 Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.bam
-rw-r--r-- 1 sr320 hyak-coenv 1.9K Mar 7 05:54 Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_PE_report.txt
Within this directory Mcap_full-u1M/ I will create two subdirectories - dedup and nodedup for downstream.
Ran jobs to deal with these separately.
#!/bin/bash
## Job Name
#SBATCH --job-name=cg
## Allocation Definition
#SBATCH --account=coenv
#SBATCH --partition=coenv
## Resources
## Nodes
#SBATCH --nodes=1
## Walltime (days-hours:minutes:seconds format)
#SBATCH --time=1-00:00:00
## Memory per node
#SBATCH --mem=100G
#SBATCH --mail-type=ALL
#SBATCH --mail-user=sr320@uw.edu
## Specify the working directory for this job
#SBATCH --chdir=/gscratch/scrubbed/sr320/030520-fr01/
# Directories and programs
bismark_dir="/gscratch/srlab/programs/Bismark-0.21.0"
bowtie2_dir="/gscratch/srlab/programs/bowtie2-2.3.4.1-linux-x86_64/"
samtools="/gscratch/srlab/programs/samtools-1.9/samtools"
# reads_dir="/gscratch/srlab/strigg/data/Pgenr/FASTQS/"
# genome_folder="/gscratch/srlab/sr320/data/geoduck/v01/"
source /gscratch/srlab/programs/scripts/paths.sh
#
#${bismark_dir}/bismark_genome_preparation \
#--verbose \
#--parallel 28 \
#--path_to_aligner ${bowtie2_dir} \
#${genome_folder}
cd /gscratch/scrubbed/sr320/030520-fr01/Mcap_full-u1M/dedup/
find *.bam | \
xargs basename -s .bam | \
xargs -I{} ${bismark_dir}/deduplicate_bismark \
--bam \
--paired \
{}.bam
${bismark_dir}/bismark_methylation_extractor \
--bedGraph --counts --scaffolds \
--multicore 14 \
--buffer_size 75% \
*deduplicated.bam
# Bismark processing report
${bismark_dir}/bismark2report
#Bismark summary report
${bismark_dir}/bismark2summary
# Sort files for methylkit and IGV
find *deduplicated.bam | \
xargs basename -s .bam | \
xargs -I{} ${samtools} \
sort --threads 28 {}.bam \
-o {}.sorted.bam
# Index sorted files for IGV
# The "-@ 16" below specifies number of CPU threads to use.
find *.sorted.bam | \
xargs basename -s .sorted.bam | \
xargs -I{} ${samtools} \
index -@ 28 {}.sorted.bam
genome_folder="/gscratch/srlab/sr320/data/froger/Mcap_Genome/"
find *deduplicated.bismark.cov.gz \
| xargs basename -s deduplicated.bismark.cov.gz \
| xargs -I{} ${bismark_dir}/coverage2cytosine \
--genome_folder ${genome_folder} \
-o {} \
--merge_CpG \
--zero_based \
{}deduplicated.bismark.cov.gz
#creating bedgraphs post merge
for f in *merged_CpG_evidence.cov
do
STEM=$(basename "${f}" .CpG_report.merged_CpG_evidence.cov)
cat "${f}" | awk -F $'\t' 'BEGIN {OFS = FS} {if ($5+$6 >= 10) {print $1, $2, $3, $4}}' \
> "${STEM}"_10x.bedgraph
done
for f in *merged_CpG_evidence.cov
do
STEM=$(basename "${f}" .CpG_report.merged_CpG_evidence.cov)
cat "${f}" | awk -F $'\t' 'BEGIN {OFS = FS} {if ($5+$6 >= 5) {print $1, $2, $3, $4}}' \
> "${STEM}"_5x.bedgraph
done
#creating tab files with raw count for glms
for f in *merged_CpG_evidence.cov
do
STEM=$(basename "${f}" .CpG_report.merged_CpG_evidence.cov)
cat "${f}" | awk -F $'\t' 'BEGIN {OFS = FS} {if ($5+$6 >= 10) {print $1, $2, $3, $4, $5, $6}}' \
> "${STEM}"_10x.tab
done
for f in *merged_CpG_evidence.cov
do
STEM=$(basename "${f}" .CpG_report.merged_CpG_evidence.cov)
cat "${f}" | awk -F $'\t' 'BEGIN {OFS = FS} {if ($5+$6 >= 5) {print $1, $2, $3, $4, $5, $6}}' \
> "${STEM}"_5x.tab
done
[sr320@mox2 dedup]$ ls
bismark_summary_report.html
bismark_summary_report.txt
CHG_CTOB_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOB_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOB_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOB_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOB_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOB_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOT_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOT_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOT_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOT_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOT_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_CTOT_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OB_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OB_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OB_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OB_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OB_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OB_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OT_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OT_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OT_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OT_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OT_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CHG_OT_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOB_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOB_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOB_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOB_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOB_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOB_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOT_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOT_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOT_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOT_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOT_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_CTOT_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OB_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OB_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OB_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OB_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OB_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OB_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OT_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OT_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OT_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OT_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OT_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CHH_OT_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOB_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOB_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOB_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOB_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOB_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOB_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOT_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOT_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOT_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOT_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOT_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_CTOT_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OB_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OB_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OB_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OB_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OB_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OB_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OT_Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OT_Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OT_Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OT_Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OT_Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.txt
CpG_OT_Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.txt
*merged_CpG_evidence.cov_10x.bedgraph
*merged_CpG_evidence.cov_10x.tab
*merged_CpG_evidence.cov_5x.bedgraph
*merged_CpG_evidence.cov_5x.tab
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe._10x.bedgraph
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe._10x.tab
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe._5x.bedgraph
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe._5x.tab
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.bam
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe..CpG_report.merged_CpG_evidence.cov
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe..CpG_report.txt
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.bam
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.bedGraph.gz
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.bismark.cov.gz
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.M-bias.txt
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.sorted.bam
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated.sorted.bam.bai
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplicated_splitting_report.txt
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_pe.deduplication_report.txt
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_PE_report.html
Meth10.fastp-trim.202003063900_R1_001_bismark_bt2_PE_report.txt
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe._10x.bedgraph
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe._10x.tab
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe._5x.bedgraph
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe._5x.tab
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.bam
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe..CpG_report.merged_CpG_evidence.cov
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe..CpG_report.txt
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.bam
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.bedGraph.gz
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.bismark.cov.gz
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.M-bias.txt
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.sorted.bam
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated.sorted.bam.bai
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplicated_splitting_report.txt
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_pe.deduplication_report.txt
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_PE_report.html
Meth11.fastp-trim.202003065734_R1_001_bismark_bt2_PE_report.txt
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe._10x.bedgraph
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe._10x.tab
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe._5x.bedgraph
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe._5x.tab
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.bam
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe..CpG_report.merged_CpG_evidence.cov
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe..CpG_report.txt
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.bam
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.bedGraph.gz
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.bismark.cov.gz
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.M-bias.txt
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.sorted.bam
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated.sorted.bam.bai
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplicated_splitting_report.txt
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_pe.deduplication_report.txt
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_PE_report.html
Meth12.fastp-trim.202003060645_R1_001_bismark_bt2_PE_report.txt
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe._10x.bedgraph
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe._10x.tab
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe._5x.bedgraph
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe._5x.tab
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.bam
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe..CpG_report.merged_CpG_evidence.cov
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe..CpG_report.txt
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.bam
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.bedGraph.gz
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.bismark.cov.gz
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.M-bias.txt
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.sorted.bam
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated.sorted.bam.bai
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplicated_splitting_report.txt
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_pe.deduplication_report.txt
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_PE_report.html
Meth16.fastp-trim.202003062412_R1_001_bismark_bt2_PE_report.txt
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe._10x.bedgraph
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe._10x.tab
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe._5x.bedgraph
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe._5x.tab
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.bam
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe..CpG_report.merged_CpG_evidence.cov
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe..CpG_report.txt
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.bam
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.bedGraph.gz
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.bismark.cov.gz
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.M-bias.txt
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.sorted.bam
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated.sorted.bam.bai
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplicated_splitting_report.txt
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_pe.deduplication_report.txt
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_PE_report.html
Meth17.fastp-trim.202003063731_R1_001_bismark_bt2_PE_report.txt
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe._10x.bedgraph
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe._10x.tab
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe._5x.bedgraph
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe._5x.tab
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.bam
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe..CpG_report.merged_CpG_evidence.cov
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe..CpG_report.txt
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.bam
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.bedGraph.gz
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.bismark.cov.gz
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.M-bias.txt
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.sorted.bam
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated.sorted.bam.bai
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplicated_splitting_report.txt
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_pe.deduplication_report.txt
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_PE_report.html
Meth18.fastp-trim.202003065117_R1_001_bismark_bt2_PE_report.txt
Yes have a .. in a few places.
https://gannet.fish.washington.edu/tmp/Mcap_full-u1M/dedup/
Same this is available for the nodedup (AKA RRBS)
https://gannet.fish.washington.edu/tmp/Mcap_full-u1M/nodedup/
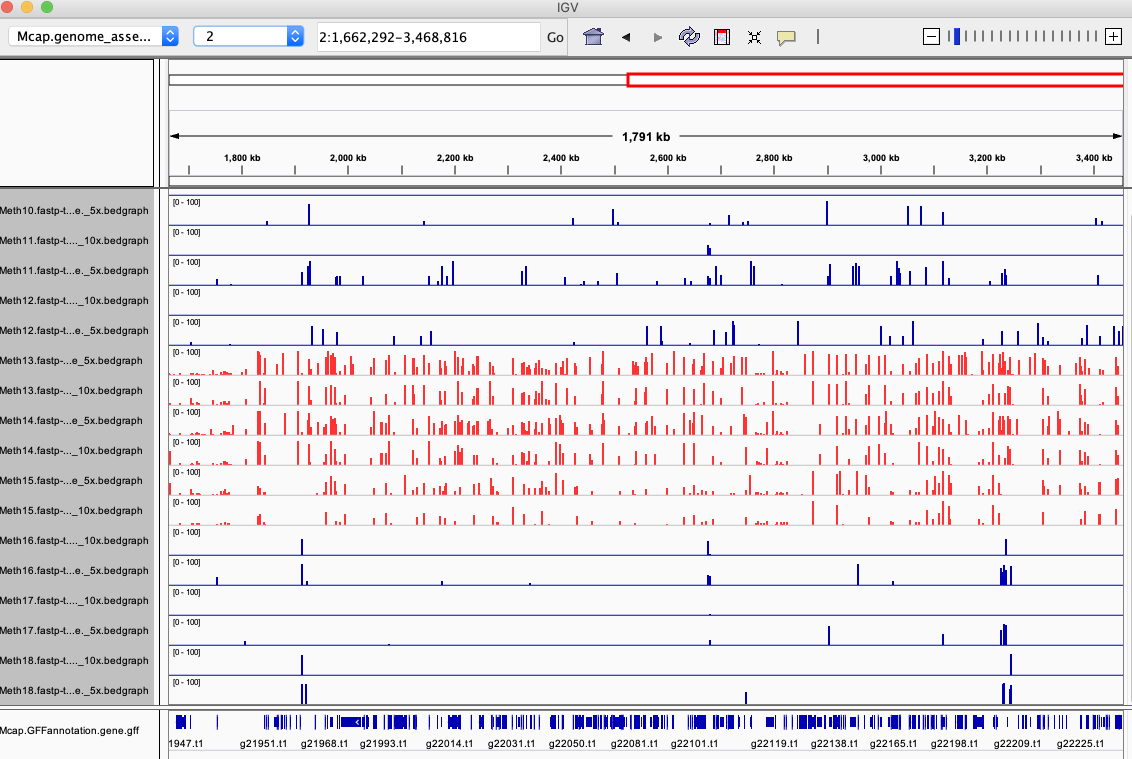
https://raw.githubusercontent.com/hputnam/Meth_Compare/master/genome-feature-files/0307_1400-igv_session.xml
Written on March 7, 2020
